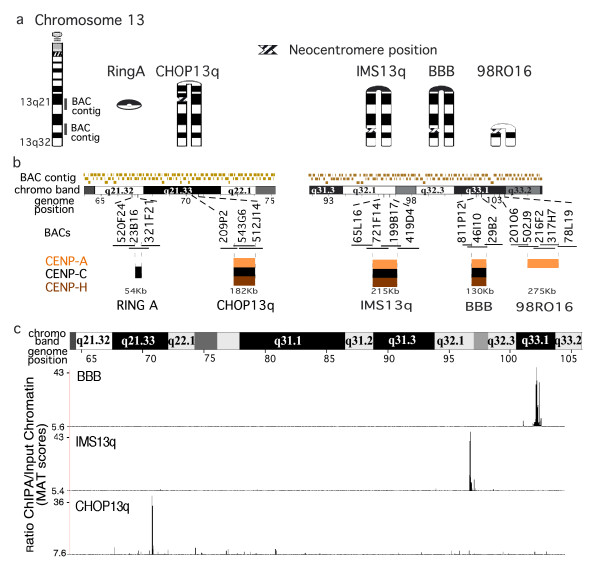
Figure 1.
Chromatin immunoprecipitation (ChIP)-CHIP (microarray) analysis of chromosome 13q neocentromeres. (a) Ideograms of four inverted duplication 13q chromosomes, shown arched at the inversion breakpoint to indicate the duplicate regions homologous to (left) the normal chromosome 13. A ring chromosome derived from band 13q21 is also shown. The name of the cell line and the cytological position of each of the neocentromeres are indicated. The position of the 13q21 and 13q32 BAC microarrays are shown on the ideogram of the normal chromosome 13. (b) Expanded areas showing the 13q21 bacterial artificial chromosome (BAC) microarray (11 Mbp, 103 BACs)[12] and the 13q32 BAC microarray (14 Mbp, 126 BACs) [11]. The region that contained each neocentromere is expanded below, showing the BACs and their overlaps in each region. The positions of centromere protein (CENP)-A, -C and/or -H are shown when determined. The results showing colocalization of CENP-A, -C and -H on the IMS13q neocentromere are modified from a previous report [12]. (c) Affymetrix high density tiling array analysis of cell lines (CHOP13q, IMS13q and BBB), showing ~42 Mbp region encompassing the BAC microarrays and region between them on chromosome 13q21 to 13q33. The distinct and specific CENP-A domain identified for each neocentromere are shown. CENP-A chromatin immunoprecipitation (ChIP) model-based analysis of tiling-array (MAT) score: CHOP13q p < 10-10; IMS13q, P < 10-8; BBB, P < 10-8).