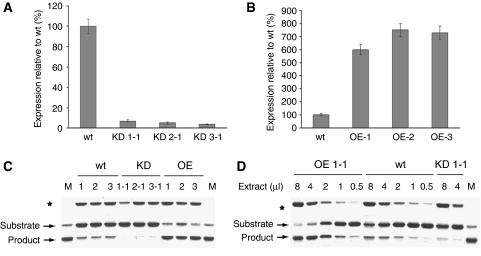
Figure 3.
Relative expression of phytaspase ORF in transgenic N. tabacum Samsun NN plants with silenced (knocked down, KD) or overexpressed (OE) phytaspase ORF. (A, B) Phytaspase ORF expression was measured by qRT–PCR in wild-type (wt) plants and in three independent phytaspase-silenced transgenic lines KD1-1, KD2-1 and KD3-1 generated using different RNAi constructs (A) and three independent lines overexpressing phytaspase (B, OE-1, OE-2 and OE-3). The results presented in (A) and (B) were typical of all transgenic lines produced. Data are means±s.d. from six independent plants. The levels of ubiquitin mRNA used as a constitutively expressed internal control were similar in all the samples analysed above (data not shown). (C, D) Transgenic tobacco plants with overexpressed (OE) and silenced (KD) phytaspase display altered phytaspase enzymatic activity relative to wild-type (wt) plants. (C) Crude leaf extracts from three independent OE and KD lines and from wt plants were tested for in vitro cleavage of GFP-VirD2Ct substrate protein. Reaction mixtures were fractionated by 12% SDS–gel electrophoresis. An unknown protein band marked with an asterisk comes from extracts and indicates similar sample loading. Lane M, substrate protein cleaved with purified tobacco phytaspase. (D) Serial dilutions of arbitrary chosen extracts of each type allow quantification of phytaspase enzymatic activity: approximately four-fold increase in OE plants and approximately eight-fold decrease in KD plants, relative to wt plants.