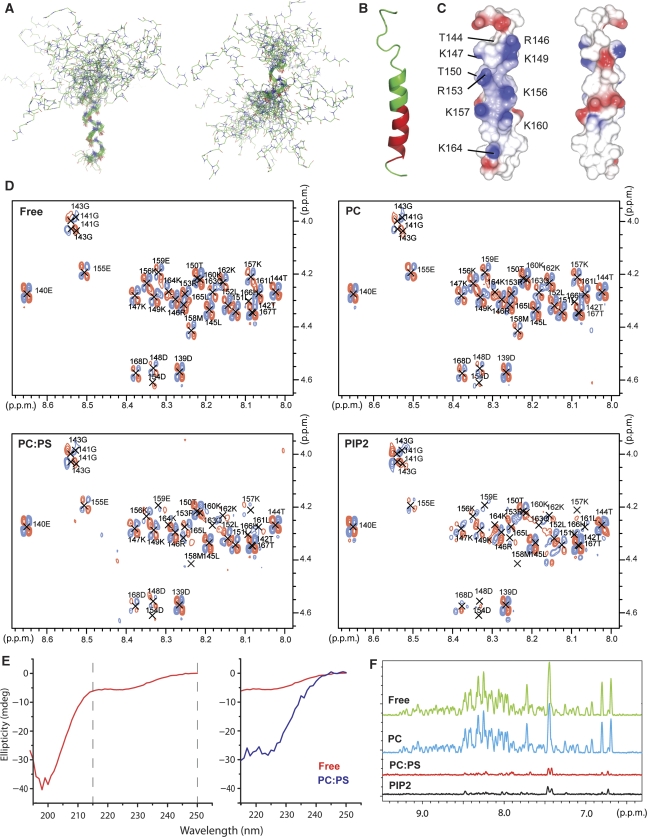
Figure 4.
Structure of the F1 loop and its function in membrane binding. (A) Superposition of the 20 lowest energy structures calculated for the F1-loop peptide in 35% TFE. Superposition was performed separately on the C-terminal helix (residues 154–167, left) and the N-terminal helix (residues 144–152, right). The middle part of the helix is not well defined in the ensemble of the calculated structure because of its lower stability, introducing a variable kink in the peptide structure. (B) Ribbon diagram of a low energy structure of the F1-loop peptide in 35% TFE corresponding to a continuous helix. Residues experiencing resonance broadening on the addition of 1:4 POPS:POPC SUVs are highlighted in red. (C) Surface charge distribution for the structure in (B). The views differ by a 180° rotation and are optimized to show the positively charged face. (D) HN/Hα region of the 2D DQF-COSY spectra of the loop peptide alone (Free) and in the presence of 2 mM SUVs composed of POPC (PC), 1:4 POPS:POPC (PC:PS) and 1:19 PIP2:POPC (PIP2) SUVs illustrating resonance broadening on addition of negatively charged lipids. (E) Secondary structure analysis of the F1-loop (145–168) peptide by circular dichroism (CD). Left—free peptide; right—superposition of the CD spectra of the free peptide (red) and the peptide in the presence of 1:4 POPS:POPC SUVs (blue) in the 215–250 nm region. Strong contribution from the SUVs to the spectrum below 215 nm prevents the detection of the peptide signal. (F) Superposition of 1H projections of F1 HSQC spectra illustrating the reduction in resonance intensities in the presence of POPS (red) and PIP2 (black), but not pure POPC (blue).