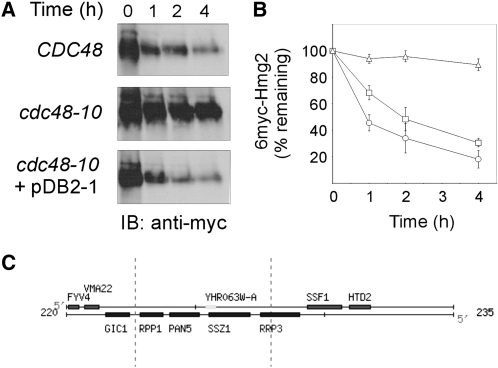
Figure 1.—
Plasmid pDB2-1, identified in a genetic screen, restores ERAD-M in cdc48-10 mutant. Turnover of 6myc-Hmg2 was measured in wild-type CDC48 (KFY100; circles), cdc48-10 (KFY194; triangles), and cdc48-10 expressing plasmid pDB2-1 identified in the genetic screen (squares). The protein 6myc-Hmg2 was expressed from pER244. (A) Following a 90-min preincubation at 37°, cycloheximide (150 μg/ml) was added and cells further incubated at 37° were collected at the indicated time points. Cells (3 A600) were lysed, total cellular proteins were resolved by SDS-PAGE, electroblotted and probed (IB) with a mouse anti-myc antibody followed by HRP-conjugated anti-mouse IgG, and HRP was visualized by ECL. (B) Blots represented by A were quantified by densitometry and 6myc-Hmg2 decay was plotted. The remaining 6myc-Hmg2 was calculated as the percentage of its level at the time of cycloheximide addition (100%). Values shown are means ± SEM of at least five independent experiments. (C) A genetic map of the insert in plasmid pDB2-1 from the Saccharomyces Genome Database (http://www.yeastgenome.org).