A mitochondrial ATP binding cassette transporter in Arabidopsis is found to play an important role in molybdenum cofactor biosynthesis. This transporter is involved in exporting cyclic pyranopterin monophosphate from the mitochondrion to the cytosol, where it is converted to molybdenum cofactor in a three-step process.
Abstract
The molybdenum cofactor (Moco) is a prosthetic group required by a number of enzymes, such as nitrate reductase, sulfite oxidase, xanthine dehydrogenase, and aldehyde oxidase. Its biosynthesis in eukaryotes can be divided into four steps, of which the last three are proposed to occur in the cytosol. Here, we report that the mitochondrial ABC transporter ATM3, previously implicated in the maturation of extramitochondrial iron-sulfur proteins, has a crucial role also in Moco biosynthesis. In ATM3 insertion mutants of Arabidopsis thaliana, the activities of nitrate reductase and sulfite oxidase were decreased to ∼50%, whereas the activities of xanthine dehydrogenase and aldehyde oxidase, whose activities also depend on iron-sulfur clusters, were virtually undetectable. Moreover, atm3 mutants accumulated cyclic pyranopterin monophosphate, the first intermediate of Moco biosynthesis, but showed decreased amounts of Moco. Specific antibodies against the Moco biosynthesis proteins CNX2 and CNX3 showed that the first step of Moco biosynthesis is localized in the mitochondrial matrix. Together with the observation that cyclic pyranopterin monophosphate accumulated in purified mitochondria, particularly in atm3 mutants, our data suggest that mitochondria and the ABC transporter ATM3 have a novel role in the biosynthesis of Moco.
INTRODUCTION
The transition element molybdenum is essential for most biological systems as it is required by a number of enzymes that catalyze diverse key reactions in the global carbon, sulfur, and nitrogen metabolism. The metal itself is biologically inactive unless it is complexed by a special cofactor. With the exception of bacterial nitrogenase, where molybdenum is a constituent of the FeMo cofactor (Dos Santos et al., 2004), molybdenum is bound to a pterin, thus forming the molybdenum cofactor (Moco), which forms the active site of all other molybdenum enzymes (Mo enzymes). In eukaryotes, the most prominent Mo enzymes are xanthine dehydrogenase (XDH), aldehyde oxidase (AO), sulfite oxidase (SO), and nitrate reductase (NR) (Mendel and Bittner, 2006). In both animals and plants, XDH is a key enzyme in purine catabolism, where it catalyzes the oxidation of hypoxanthine to xanthine and of xanthine to uric acid. Plant and animal SO proteins are involved in the detoxification of excess sulfite and the degradation of sulfur-containing amino acids, whereas AO proteins catalyze the oxidation of a variety of aldehydes. While the in vivo function of animal AO proteins is still relatively unclear (Garattini et al., 2008), the physiological role of plant AO proteins is much better understood. Arabidopsis aldehyde oxidase 3 (AAO3), one of the AO isoenzymes from Arabidopsis thaliana, catalyzes the oxidation of abscisic aldehyde to the phytohormone abscisic acid (ABA) (Seo et al., 2000a, 2000b), which is required for seed and plant development, stomatal regulation, gene regulation, and response to a variety of abiotic stresses (summarized in Seo and Koshiba, 2002; Verslues and Zhu, 2005). NR, which is exclusively present in autotrophic organisms like plants, algae, yeasts, and fungi, catalyzes the key step in inorganic nitrogen assimilation, namely, the reduction of nitrate to nitrite. Recently, a novel Mo enzyme has been identified in the outer membrane of pig liver mitochondria (Havemeyer et al., 2006). Although the enzyme was shown to be capable of catalyzing the N reduction of amidoximes, its physiological function is as yet unknown.
The biosynthesis of Moco, the prosthestic group common to all aforementioned enzymes, can be devided into four steps, which require the direct action of six proteins. In the first step, 5′-GTP is converted to precursor Z (Haenzelmann and Schindelin, 2004), which was recently renamed cyclic pyranopterin monophosphate (cPMP) (Schwarz, 2005). This reaction is catalyzed by two proteins, the cofactor for nitrate reductase and xanthine dehydrogenase (CNX) proteins CNX2 and CNX3 in plants (Hoff et al., 1995) and the molybdenum cofactor synthesis (MOCS) proteins MOCS1A and MOCS1B in humans (Haenzelmann et al., 2002). CNX2 and MOCS1A belong to the superfamily of S-adenosylmethionine–dependent radical enzymes, members of which catalyze the formation of protein and/or substrate radicals by reductive cleavage of S-adenosylmethionine by a protein-bound [4Fe-4S] cluster. In fact, MOCS1A was shown to be capable of binding two highly oxygen-sensitive iron-sulfur (Fe-S) clusters of the [4Fe-4S] type, an NH2-terminal cluster essential for activity, and a C-terminal cluster required for proper folding of the protein (Haenzelmann et al., 2004). The precise functions of CNX3 and MOCS1B are not presently clear, but since some of the S-adenosylmethionine–dependent radical enzymes require another protein onto which the radical is transferred, it is speculated that CNX3 and MOCS1B might have a similar function. In step 2 of Moco biosynthesis, a dithiolene group is introduced into cPMP by transfer of two sulfur atoms, resulting in the formation of molybdopterin (MPT). This reaction is catalyzed by the enzyme MPT synthase, a heterotetrameric complex of two large and two small subunits (CNX6 and CNX7 in plants and MOCS2A and MOCS2B in humans, respectively) that stoichiometrically converts cPMP into MPT. After MPT synthase has transferred the two sulfurs to cPMP, it has to be resulfurized by the MPT-synthase sulfurylase. This resulfuration is catalyzed by plant CNX5 or human MOCS3 and involves the adenylation of MPT synthase followed by sulfur transfer (Matthies et al., 2004, 2005). In step 3, MPT is bound by the two-domain proteins, CNX1 in plants and GEPHYRIN in humans, and is activated by adenylation, whereby MPT-AMP is formed (Kuper et al., 2004). This reaction is restricted to the G-domains of these proteins and is assumed to prepare MPT for insertion of molybdenum. During the fourth and final step, MPT-AMP is transferred to the E-domains of CNX1 and GEPHYRIN, respectively, where the adenylate is cleaved and molybdenum is inserted to finally yield mature Moco (Llamas et al., 2006). While the enzymes of steps 2 to 4 (Matthies et al., 2004; Lardi-Studler et al., 2007; Gehl et al., 2009) have been found to be located in the cytosol, the subcellular localization of step 1 is unclear. Hoff et al. (1995) realized that the enzymes involved in step 1, CNX2 and CNX3, carry NH2-terminal extensions that may represent targeting signals for the import into chloroplasts or mitochondria. Yet, studies with the in vitro translated proteins and purified chloroplasts and mitochondria failed to show import of these proteins into either of these organelles (Hoff et al., 1995).
A deficiency in the Moco biosynthetic pathway results in the pleiotropic loss or reduction of all Mo enzyme activities. In humans, Moco deficiency is characterized by progressive neurological damage, in most cases leading to death in early childhood. This severe phenotype is mainly caused by the deficiency of SO, which protects the organism, in particular the brain, from accumulation of toxic sulfite (Johnson and Duran, 2001). In plants, a loss of Moco biosynthetic activity affects nitrogen assimilation and phytohormone biosynthesis, caused by the deficiencies of NR and AO, respectively. Such plants are unable to survive under normal growth conditions and can be cultivated only under precisely adopted conditions that include ammonium as an alternative nitrogen source and increased relative humidity. So far, Moco deficiency has exclusively been reported to result from a mutation in any of the genes directly involved in Moco biosynthesis. However, since CNX2 and MOCS1A require Fe-S clusters for activity, it is reasonable to assume that a Moco-deficient phenotype may also be caused by a deficiency in the biosynthesis of Fe-S clusters. In support of this, Arabidopsis lines with downregulated levels of NFS1, the mitochondrial Cys desulfurase with an essential role in Fe-S cluster biosynthesis, had strongly decreased AO activities (Frazzon et al., 2007). However, whether this is due to decreased activity of CNX2 or to Fe-S assembly on cytosolic AO proteins was not investigated.
With the exception of plastids that are equipped to synthesize Fe-S clusters for their own set of Fe-S proteins (Pilon et al., 2006), all cellular Fe-S proteins depend on the mitochondrial Fe-S cluster assembly machinery (Balk and Lobréaux, 2005; Lill and Muehlenhoff, 2008) and the ATP binding cassette (ABC) transporter in the inner membrane of mitochondria, which transports an as yet unidentified component (Lill and Muehlenhoff, 2008) from the mitochondrial matrix into the cytosol. This ABC transporter has been investigated most intensively in Saccharomyces cerevisiae, where it is referred to as Atm1p (Kispal et al., 1999). Each Atm1p protein consists of one domain that comprises six transmembrane regions that form a membrane-spanning channel and one ATP binding domain, which faces the mitochondrial matrix and is required for mobilizing energy from ATP hydrolysis for active transport of the as yet unidentified component across the inner membrane. By complementation of a yeast Atm1p-deficient mutant, which is characterized by a deficiency of cytosolic Fe-S proteins, strong growth defects and accumulation of iron in the mitochondria, functional homologs have been identified also in humans (ABCB7; Allikmets et al., 1999) and plants (Sta1/ATM3; Kushnir et al., 2001; Chen et al., 2007; Bernard et al., 2009). While a mutation in human ABCB7 causes X-linked sideroblastic anemia and cerebellar ataxia (Allikmets et al., 1999), the sta1/atm3 mutant of Arabidopsis that carries a T-DNA insertion in the ATM3 gene is characterized by dwarfism, chlorosis, altered morphology of leaves and cell nuclei (Kushnir et al., 2001), and reduced activities of cytosolic Fe-S proteins (Bernard et al., 2009). Recently, in an effort to establish a unified nomenclature, the gene symbols AtABCB24, AtABCB23, and AtABCB25 have been suggested for ATM1, ATM2, and ATM3, respectively (Verrier et al., 2008). However, for consistency with previous publications, we have adhered to the ATM gene symbols in this work.
Since up to now no attempts have been undertaken to investigate the crosstalk between Moco and Fe-S cluster biosynthesis, this work aims to study the importance of ATM3 for Moco biosynthesis, with particular attention to the subcellular localization of step 1. If the Fe-S protein CNX2, which is involved in the first step of Moco biosynthesis, is located in the cytosol, it must depend on the function of ATM3. Thus, a deficiency in ATM3 should affect CNX2 activity, thereby resulting in diminished production of cPMP and of all further downstream Moco intermediates. If, however, CNX2 and the entire step 1 of Moco biosynthesis is located in mitochondria, as suggested by Hoff et al. (1995), CNX2 activity would be independent of ATM3 functionality. Here, we demonstrate that a mutation in Arabidopsis ATM3 leads to decreased levels of Moco, but to a two- to threefold increase of cPMP levels. Moreover, the increase of cPMP in the ATM3-deficient plants was much more prominent in the purified mitochondria. Together with immunolocalization data showing that the proteins involved in step 1 of Moco biosynthesis, CNX2 and CNX3, are localized in mitochondria, we suggest that step 1 of Moco biosynthesis, the conversion of 5′-GTP to cPMP, occurs in mitochondria rather than in the cytosol. As the accumulation of cPMP in mitochondria seems to be a direct consequence of the ATM3 mutation, our data indicate that ATM3 is involved in the export of cPMP from mitochondria into the cytosol. These findings provide evidence for a novel function of mitochondria and the mitochondrial ABC half-transporter ATM3 in Moco biosynthesis.
RESULTS
Mo Enzyme Activities and Protein Levels Are Reduced in atm3 Mutants
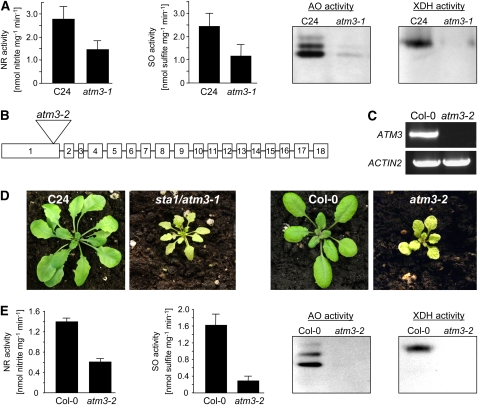
The Arabidopsis starik1(sta1) mutant, which carries a T-DNA insertion in the ATM3 gene (Babiychuk et al., 1997; Kushnir et al., 2001) and which we renamed atm3-1, was investigated with respect to the influence of the mutation in ATM3 on Moco homeostasis. As described by Kushnir et al. (2001), homozygous atm3-1 mutants exhibited symptoms of chlorosis and altered leaf morphology and, relative to the corresponding wild-type C24, were significantly smaller in size when grown in potting soil. To study whether, besides its presumed effect on the maturation of extramitochondrial Fe-S proteins (Kushnir et al., 2001; Bernard et al., 2009), the mutation in ATM3 also directly or indirectly affects Moco-dependent proteins, the activities of the Mo enzymes NR, SO, AO, and XDH were analyzed. While NR and SO activities were reduced by ∼50% in leaf extracts of atm3-1 mutants, the activities of both XDH and AO were nearly fully abolished (Figure 1A), thereby suggesting an important role for ATM3 in Mo enzyme maturation.
Figure 1.
Phenotypical Characterization of atm3-1 and atm3-2 Mutants.
(A) Mo enzyme activities (NR, SO, AO, and XDH) in leaves of the sta1/atm3-1 mutant and the C24 wild type (NR, n = 4; SO, n = 6; bars represent se of the means). For comparison of AO and XDH activities, leaf extracts were electrophoresed on native polyacrylamide gels and either stained for AO activity or for XDH activity. For each, one representative gel of six experiments is shown.
(B) T-DNA insertion site in the Arabidopsis GABI-Kat atm3-2 mutant.
(C) RT-PCR–based detection of full-length ATM3 trancripts in Columbia-0 (Col-0) wild types and atm3-2 mutant plants (n = 3). Actin2 primers were used to demonstrate successful reverse transcription of mRNA.
(D) Phenotypical comparison of atm3-1 and atm3-2 mutants.
(E) Mo enzyme activities (NR, SO, AO, and XDH) in leaves of the GABI-Kat atm3-2 mutant and the Col-0 wild type (NR, n = 4; SO, n = 3; bars represent seof the means). For AO and XDH activities, one representative gel of four experiments is shown.
To confirm these results, a new T-DNA insertion mutant allelic to sta1/atm3-1 was isolated. From segregating T3 lines of the GABI-Kat collection (GK-714C03.01) we obtained plants that were homozygous for a T-DNA insertion in ATM3. Sequence analysis of the T-DNA flanking regions indicated that the T-DNA is located between bases 422 and 437 following the start ATG in exon 1 of ATM3 (Figure 1B). RT-PCR analysis failed to detect full-length ATM3 transcripts in this mutant (Figure 1C), suggesting a severe effect on the expression of ATM3 protein. Like the atm3-1 mutant, the genome of the newly isolated atm3 mutant did not contain any other T-DNA insertion, as suggested by segregation analysis and inverse PCR. According to our nomenclature, the newly identified GABI-Kat mutant consequently was renamed into atm3-2.
When growing side by side, 5-week-old atm3-1 and atm3-2 mutants that were homozygous for the insertion in ATM3 showed nearly identical phenotypes (Figure 1D). In contrast with atm3-1 mutants, however, the mutation in homozygous atm3-2 plants lead to male sterility; thus, atm3-2 mutants were unable to produce viable seeds. Each new generation of homozygous atm3-2 mutants therefore had to be selected from segregating heterozygous atm3-2 plants. When analyzing leaf extracts of atm3-2 mutant plants for Mo enzyme activities, nearly identical results as observed for the atm3-1 mutant were obtained (Figure 1E), demonstrating that the reduction of Mo enzyme activities in both mutants is a direct consequence of the respective T-DNA insertion in the ATM3 gene. It should be noted that in both mutants the activities of XDH and AO, which besides Moco and FAD require Fe-S clusters as prosthetic groups for enzymatic activity, were affected more strongly than those of NR and SO. This may be indicative of XDH and AO being affected by a severe ATM3-related limitation of cytosolic Fe-S clusters in combination with Moco deficiency, while the effect on NR and SO in the atm3 mutants is solely derived from a deficiency in Moco metabolism.
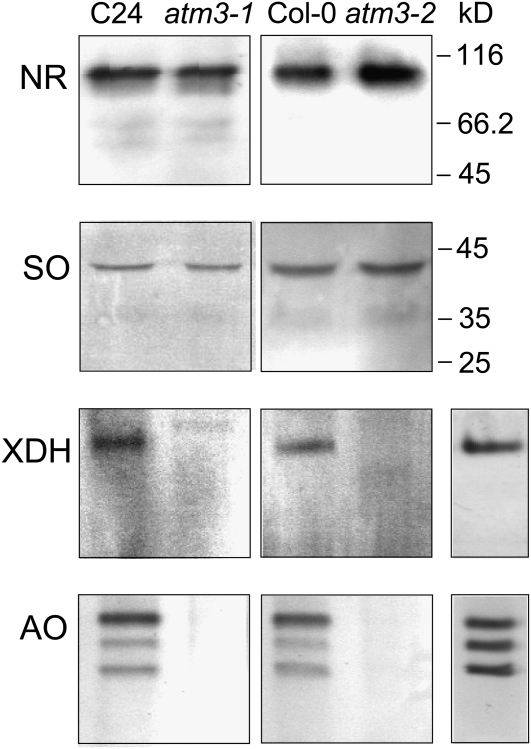
To investigate whether the reduction in Mo enzyme activities is reflected at the protein level, immunoanalysis was performed using leaf extracts of atm3-1 and atm3-2 mutants. Although in both mutants the activities of NR and SO were reduced, the total levels of NR and SO proteins appeared to be unaltered (Figure 2). In contrast with this, no XDH and AO proteins could be detected, which is consistent with the strong reduction of XDH and AO activies and indicates that the insertion of Fe-S clusters into XDH and AO is a prerequisite for proper maturation and stability of these proteins. This assumption is confirmed by our finding that mutants with a deficiency in the Moco biosynthetic pathway, like the mutants sir5/cnx7, sir/cnx1 (Dai et al., 2005), and chl5 (Crawford, 1992), display unchanged protein levels (see Supplemental Figure 1 online).
Figure 2.
Immunodetection of Molybo Enzymes in atm3-1 and atm3-2 Mutants.
Total extracts from leaves of atm3-1 and atm3-2 mutants and their respective wild types (C24 and Col-0, respectively) were analyzed for their Mo enzyme contents using anti-NR antibodies, anti-SO antibodies, anti-XDH antibodies, or anti-AAO1 antibodies. XDH and AO activity staining (single lanes in right-most panel) of the same native polyacrylamide gels as used for immunoanalysis served as markers for XDH and AO proteins, respectively.
atm3-1 Mutants Show Altered ABA Response upon Drought Stress
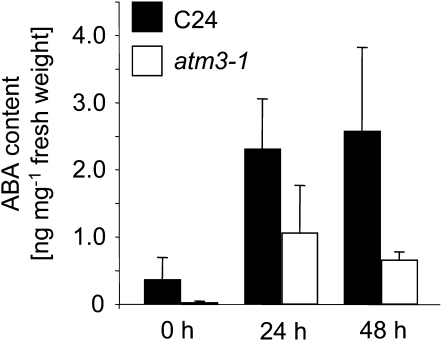
The Arabidopsis genome harbors four AO genes, AAO1 to AAO4, whose products form homodimers and heterodimers, thereby leading to altered substrate specificities of the respective isoenzymes. In seedlings, the proteins AAO1 and AAO2 form three homo- and heterodimeric AO isoenzymes (Akaba et al., 1999), while in rosette leaves, AAO1 proteins are replaced by AAO3 proteins, thereby resulting in a different composition of homo- and heterodimeric isoforms (Seo et al., 2000a). One of these isoforms, the AAO3 homodimer, catalyzes the last step of ABA biosynthesis, the oxidation of abscisic aldehyde to ABA (Seo et al., 2000a, 2000b). In contrast with AAO1-3, AAO4 appears to be present in siliques only (Ibdah et al., 2009). Since AO activities and protein levels were markedly reduced in rosette leaves of the atm3 mutants, the phenotype of these mutants may also be linked to ABA deficiency. Thus, we determined the ability of atm3-1 mutants to produce ABA under both standard growth and drought stress conditions, the latter of which typically requires enhanced levels of ABA for a proper stress response. As shown in Figure 3, atm3-1 plants already accumulated less ABA than the respective wild-type plants under standard growth conditions. When the plants were exposed to drought stress, atm3-1 mutants were capable of sythesizing more ABA. Yet, relative to the respective wild types, the drought-dependent accumulation of ABA was significantly less pronounced in mutant plants, indicating that the reduction of AO proteins and activities in atm3 mutants is indeed accompanied by a reduced ability to produce ABA under standard conditions and in response to drought stress.
Figure 3.
ABA Contents in atm3-1 Mutants after Drought Stress.
For drought stress treatment, atm3-1 mutants and Col-0 wild types were removed from the soil and placed in covered Petri dishes for the indicated periods (n = 6; bars represent se of the means).
MPT/Moco Levels Are Reduced, but cPMP Levels Are Enhanced, in Leaf Extracts of atm3 Mutants
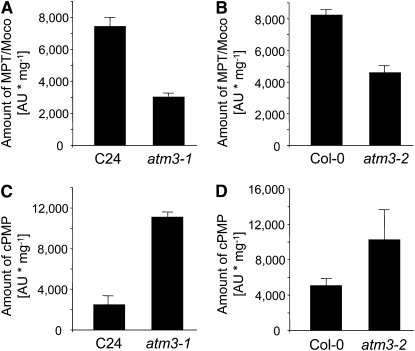
CNX2, one of the enzymes involved in the first step of Moco biosynthesis in Arabidopsis, is an ortholog of human MOCS1A and Escherichia coli MoaA, which both belong to the S-adenosylmethionine–dependent radical enzyme superfamily and depend on two [4Fe-4S] clusters for catalyzing the conversion of 5′-GTP to cPMP (Haenzelmann et al., 2004; Haenzelmann and Schindelin, 2004). Since previous attempts to localize CNX2 and its partner protein CNX3 to mitochondria or chloroplasts failed (Hoff et al., 1995), likely because some unknown component required for import of CNX2 and CNX3 was lacking in these in vitro experiments, the possibility of a cytosolic localization of these enzymes was considered. If step 1 of Moco biosynthesis were localized in the cytosol, the activity of the Fe-S enzyme CNX2 would be directly affected by a mutation in ATM3, as was observed for the cytosolic isoform of aconitase, a [4Fe-4S] enzyme (Bernard et al., 2009). A lack of extramitochondrial Fe-S clusters would not allow proper assembly of holo-CNX2 and thus would result in reduced or no CNX2 activity, accompanied by reduced amounts of cPMP and all downstream derivatives. Consistent with these expectations, the levels of MPT and Moco, measured as their common oxidation product FormA dephospho, were found to be decreased in leaf extracts of both atm3 mutants (Figures 4A and 4B). The total amount of cPMP, however, which represents the first stable intermediate of Moco biosynthesis and the direct product of CNX2 and CNX3, was significantly increased rather than decreased in both atm3 mutants (Figures 4C and 4D). According to these findings, CNX2 must be active and fully constituted with Fe-S clusters in the atm3 mutants, indicating that it does not depend on the function of mitochondrial ATM3. This result casts doubt on an extramitochondrial localization of the first step of Moco biosynthesis. The observation that the expression levels of none of the CNX genes were significantly altered in the atm3 mutants (see Supplemental Figure 2 online) also makes regulatory effects, caused by the ATM3 deficiency, on Moco biosynthesis unlikely.
Figure 4.
Amounts of Moco Intermediates in Leaves of atm3-1 and atm3-2 Mutants.
MPT/Moco amounts in leaf extracts of atm3-1 (A) and atm3-2 (B) mutants were measured as FormA dephospho. cPMP amounts in leaf extracts of atm3-1 (C) and atm3-2 (D) mutants were measured as CompoundZ. Data represent averages from four experiments. Bars represent se of the means.
The First Step of Moco Biosynthesis Is Localized in Mitochondria
The analysis of cPMP and MPT/Moco in leaf extracts of the atm3 mutants suggested that the first step of Moco biosynthesis, the CNX2/CNX3-catalyzed conversion of 5′-GTP into cPMP, is not affected by a deficiency in ATM3. As CNX2 activity is preserved in atm3 mutants, CNX2 is rather present in the holo-[4Fe-4S]2-form and thus is likely to be localized in mitochondria where cellular Fe-S clusters, except for plastidic Fe-S proteins, are originally synthesized. A mitochondrial localization of CNX2, and of CNX3, is also suggested by the presence of NH2-terminal extensions that are predicted to represent mitochondrial import signals (Subcellular Proteomic Database).
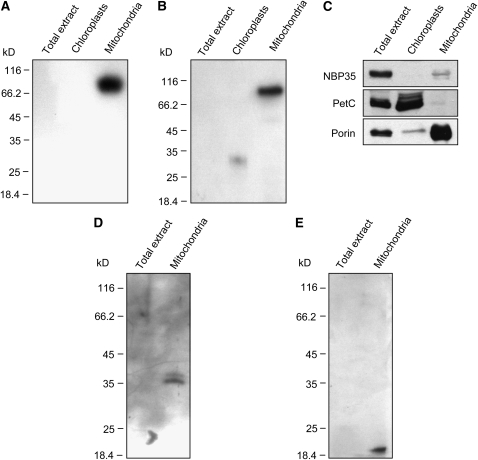
To further investigate the localization of CNX2 and CNX3, mitochondria were isolated from Arabidopsis leaves and subjected to immunoanalysis using antibodies specifically raised against recombinant CNX2 and CNX3 proteins. As mitochondrial import signals are difficult to discriminate from chloroplast import signals, isolated chloroplasts were likewise used for CNX2/CNX3 immunostudies. After standard SDS-PAGE and immunoblotting, both anti-CNX2 and anti-CNX3 antibodies detected cross-reacting proteins in Arabidopsis mitochondria but failed to detect any proteins in chloroplasts or total extracts (Figures 5A to 5C). These findings demonstrate that CNX2 and CNX3 are indeed localized in mitochondria and not in chloroplasts. The observation that no proteins were detected in total extracts indicates a low abundance of CNX2 and CNX3 proteins (e.g., in comparison to the highly abundant porin) (Figure 5C), which is consistent in particular with a very low abundance of CNX3 mRNA (see Supplemental Figure 2 online; Hoff et al., 1995). Interestingly, when immunoanalysis was performed after standard SDS-PAGE, which included the presence of mercapthoethanol and boiling for 10 min, the molecular weight of the cross-reacting proteins did not correspond to the calculated masses of mature CNX2 and CNX3 monomers (∼36 and ∼18 kD, respectively) but to the calculated masses of CNX2 dimers and CNX3 hexamers (∼72 and ∼108 kD, respectively). These findings are in good agreement with the three-dimensional structures of the bacterial orthologs of CNX2 and CNX3, MoaA and MoaC, which demonstrated that MoaA and MoaC likewise form dimers (Haenzelmann and Schindelin, 2004) and hexamers (Wuebbens et al., 2000), respectively. When SDS-PAGE was performed in the presence of iodoacetamide, a strong alkylating sulfhydryl reagent that is commonly used to prevent disulfide bond formation, only bands corresponding to CNX2 and CNX3 monomers were detectable (Figures 5D and 5E), thereby supporting the identity of these bands as CNX2 and CNX3 proteins.
Figure 5.
Immunodetection of CNX2 and CNX3 in Mitochondria of Wild-Type Plants.
Using specific anti-CNX2 (A) and anti-CNX3 (B) antibodies, cross-reacting proteins were detected exclusively in mitochondria. The purity of the fractions was confirmed by probing the same membrane for cytosolic NBP35 protein, chloroplastidic PetC, and mitochondrial Porin (C). In the presence of iodoacetamide, anti-CNX2 (D) and anti-CNX3 (E) antibodies recognized proteins of ∼36 and ∼18 kD, respectively, that correspond to the masses of mature CNX2 and CNX3 monomers without their respective NH2-terminal signal sequences.
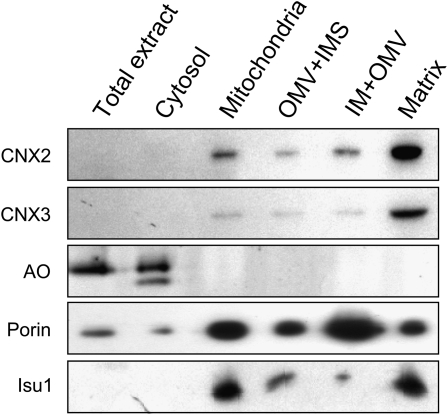
For a more detailed analysis of the localization of CNX2 and CNX3, we prepared mitochondria from cauliflower (Brassica oleracea), which is closely related to Arabidopsis but, in contrast with Arabidopsis, allows for the preparation of large quantities of mitochondria from chloroplast-free tissues. The obtained mitochondria were further subfractionated into three fractions consisting of (1) outer membrane vesicles (OMVs) and proteins of the intermembrane space, (2) a mixed fraction of OMV and the inner mitochondrial membrane, and (3) the mitochondrial matrix (Millar et al., 2001). The purity of the subfractions was verified using antibodies raised against potato (Solanum tuberosum) porin and Arabidopsis Isu1, which specifically localize to the outer membrane (Heins et al., 1994) and the matrix (Léon et al., 2005), respectively. To estimate the degree of cytosolic contaminations, the fractions were stained for the activity of AO, which is known to exclusively localize to the cytosol (Melhorn et al., 2008). As observed before in Arabidopsis extracts (Figure 5), both anti-CNX2 and anti-CNX3 antibodies exclusively cross-reacted with proteins in the mitochondrial preparations but not with those from the cytosol (Figure 6). Among the mitochondrial subfractions, anti-CNX2 and anti-CNX3 cross-reacting proteins were most abundant in the matrix fraction, suggesting that the NH2-terminal extensions of CNX2 and CNX3 indeed represent targeting sequences for the import of both proteins into the mitochondrial matrix.
Figure 6.
Localization of CNX2 and CNX3 in the Mitochondrial Matrix.
Mitochondria were subfractionated into a fraction consisting of OMVs and inner membrane space (IMS), a fraction consisting of inner membrane (IM), OMV, and the matrix fraction. All fractions (10 μ g each) were immunoanalyzed for the presence of CNX2 and CNX3 proteins using anti-CNX2 and anti-CNX3 antibodies, respectively. The purity of the mitochondrial fractions was confirmed by probing the same membrane for the mitochondrial proteins porin (outer membrane) and Isu1 (matrix). AO activity staining of these fractions allowed estimation of the degree of contamination with cytosolic proteins.
The Phenotype of Moco Mutants with a Deficiency in Step 2 of Moco Biosynthesis Differs from That of atm3 Mutants
To provide further evidence that the Moco-related phenotype of the mutants atm3-1 and atm3-2 is directly derived from a deficiency in ATM3 rather than from an unknown effect on step 2 of Moco biosynthesis, we analyzed the sir1/cnx5 mutant of Arabidopsis (Zhao et al., 2003). CNX5 represents the MPT synthase sulfurylase, which is essential for the regeneration of the MPT synthase complex that catalyzes the conversion of cPMP to MPT (Mendel and Bittner, 2006). A lack of CNX5 activity therefore is likely to result in accumulation of cPMP, similar to the accumulation observed in the atm3 mutants, raising the question of whether these two types of mutations can be biochemically discriminated.
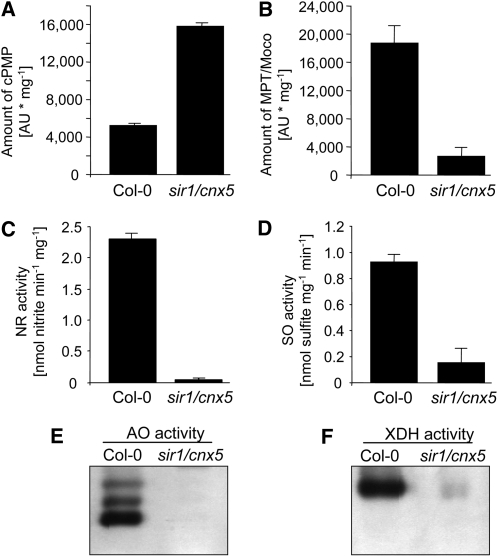
Like the mutants atm3-1 and atm3-2, the sir1/cnx5 mutant is characterized by a strong accumulation of cPMP (Figure 7A). In contrast with the atm3 mutants, however, sir1/cnx5 mutant plants do not display a moderate decrease of MPT/Moco levels, but a dramatic reduction of these molecules (Figure 7B) accompanied by strongly reduced Mo enzyme activities (Figures 7C to 7F). It is important to note that all Mo enzyme activities in extracts of sir1/cnx5 mutants were equally reduced to very low levels, whereas in extracts of atm3 mutants, only the activities of XDH and AO, but not those of NR and SO, were reduced with similar strength (Figure 1). It thus appears reasonable to state that in the sir1/cnx5 mutant all Mo enzymes are affected by the limited availability of Moco, which is in contrast with the situation in the atm3 mutants, where NR and SO are affected solely by the decrease in Moco levels, while XDH and AO appear to be affected mainly by the strong decrease in cytosolic Fe-S clusters in addition to decreased Moco levels.
Figure 7.
Biochemical Characterization of the sir1/cnx5 Mutant.
Leaf extracts of Arabidopsis sir1/cnx5 mutants with a deficiency in step 2 of Moco biosynthesis were analyzed for their cPMP contents (A) and MPT/Moco contents (B), as well as for the activities of NR (C), SO (D), AO (E), and XDH (F). cPMP and MPT/Moco were measured as CompoundZ (n = 3) and FormA dephospho (n = 3), respectively. NR and SO activities are given as mean ± sd(n = 4). For AO and XDH, activities of one representative gel of four experiments are shown.
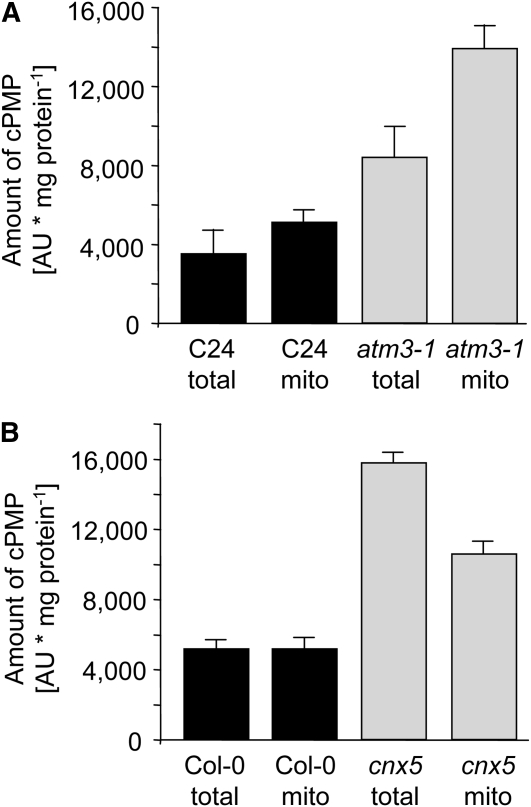
To further discriminate between atm3 and cnx5 mutants, we studied whether cPMP accumulates in the mitochondria or the cytosol of these plants. Relative to total leaf extracts, cPMP levels were found to be only slightly elevated in isolated mitochondria of C24 wild-type plants (Figure 8A). In comparison to total extracts and mitochondria of C24 wild-type plants, cPMP levels were considerably higher in total extracts of atm3-1 mutants but were by far most pronounced in isolated mitochondria of atm3-1 plants (Figure 8A). In contrast with this, the mitochondrial cPMP levels of sir1/cnx5 mutants did not exceed the levels of the respective total extract, indicating that cPMP accumulates in the cytosol rather than in mitochondria (Figure 8B). Thus, ATM3-deficient mutants are likely to be impaired in the transport of cPMP across the mitochondrial membranes but retain the capacity to convert cPMP into MPT and Moco. By contrast, CNX5-deficient mutants are able to export cPMP from mitochondria to the cytosol but have lost the ability to further process cPMP to MPT and Moco. This shows that CNX5, representing step 2 of Moco biosynthesis, functions downstream of ATM3 during Moco biosynthesis. Another possible explanation for the cPMP accumulation in atm3 mutants may be that the Moco-related function of ATM3 is to export a mitochondrial compound that serves as sulfur donor for the MPT synthase sulfurylase CNX5 in the cytosol. However, in that case, cPMP would accumulate in the cytosol and the phenotype of atm3 mutants would be nearly indistinguishable from the phenotype of cnx5 mutants. Taken together, these findings show that mutations in ATM3 and CNX5 have different effects on cPMP distribution, Moco biosynthesis, and Mo enzyme activities.
Figure 8.
Determination of cPMP in Mitochondria of atm3-1 and cnx5 Mutants.
cPMP amounts in total and mitochondrial (mito) extracts from leaves of C24 wild types and atm3-1 mutants ([A]; n = 4), and Col-0 wild types and sir1/cnx5 mutants ([B]; n = 3). Bars represent se of the means.
DISCUSSION
By use of specific antibodies raised against recombinant CNX2 and CNX3 proteins, we showed that the first step of Moco biosynthesis, the CNX2/CNX3-catalyzed conversion of 5′-GTP to cPMP, proceeds in the mitochondrial matrix (Figure 5). A mitochondrial localization of step 1 is further supported by our finding that cPMP accumulates in isolated mitochondria of atm3-1 mutants but not in mitochondria of CNX5-deficient plants (Figure 8). In fact, the localization of CNX2 and CNX3 in this particular organelle appears reasonable from a physiological point of view as the mitochondrial matrix basically is equipped to provide (1) 5′-GTP as substrate for cPMP synthesis, (2) Fe-S clusters as the essential prosthetic group for CNX2 (Balk and Lobréaux, 2005; Lill and Muehlenhoff, 2008), and (3) a reducing environment for stabilization of these oxygen-sensitive clusters bound to CNX2 (Haenzelmann et al., 2004). However, since steps 2 to 4 of Moco biosynthesis have been demonstrated to be localized in the cytosol (Matthies et al., 2004; Lardi-Studler et al., 2007; Gehl et al., 2009), export of cPMP from the mitochondria into the cytosol is required to allow further processing to MPT and Moco. In this respect, our observation that cPMP accumulates in mitochondria of ATM3-deficient plants suggests that the ABC half-transporter ATM3 represents the link between step 1, the conversion of 5′-GTP into cPMP in the mitochondrial matrix, and step 2, the conversion of cPMP into MPT in the cytosol. It therefore appears reasonable to presume that ATM3 has a function in the export of cPMP from mitochondria into the cytosol.
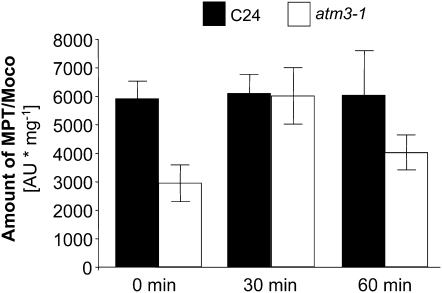
Although both atm3 mutants carry T-DNA insertions within the coding sequence of ATM3, which usually results in the total knockout of the affected gene, significant amounts of Moco (40 to 50%; Figure 4) are still synthesized in these mutants. A residual ATM3 activity could be possible in the sta1/atm3-1 mutant, as the T-DNA interrupts the coding sequence of the ATM3 mRNA after the transmembrane domain and generates a fusion construct of the ATM3 transmembrane domain with neomycin phosphotransferase brought by the T-DNA (Kushnir et al., 2001). Although the resulting chimeric protein lacks the entire ATP binding domain, passive but selective transport may be retained to some extent. In fact, little but detectable AO and XDH activity has been demonstrated for the atm3-1 mutant (Figure 1A; Bernard et al., 2009), indicating that, besides cPMP, Fe-S cluster precursors must also have been transported across the mitochondrial membrane and suggesting that ATM3 is not completely absent in this mutant. In contrast with the atm3-1 mutant, the T-DNA insertion in the atm3-2 mutant is located in exon 1 and thus impedes the correct expression not only of the ATP binding domain but even of the membrane-spanning domain. The atm3-2 mutant therefore is likely to represent a complete knockout mutant, which is confirmed by the total absence of ATM3 transcripts and sterility of homozygous plants. Nevertheless, like atm3-1 plants, atm3-2 plants also exhibit residual activity of cytosolic Fe-S proteins, such as XDH and cytosolic aconitase (Bernard et al., 2009), which shows that, besides Moco metabolism, cytosolic Fe-S cluster assembly is also retained to a small extent in this mutant. A similar observation was made for the yeast Atm1-deficient mutant, which likewise exhibited residual activity of a cytosolic Fe-S protein, Leu1 (Bernard et al., 2009), indicating that some amount of Fe-S equivalents can also be bypassed in Atm1-deficient yeast. Therefore, it appears reasonable to assume that such compensatory systems for Fe-S and for cPMP may also exist in the Arabidopsis T-DNA insertion mutants atm3-1 and atm3-2. In Arabidopsis, such a system may be represented by the ATM3 homologous transporters ATM1 and/or ATM2. Similar to ATM3, both possess the capacity to localize to mitochondria (Chen et al., 2007), which should be a prerequisite for compensation of the ATM3-deficient phenotype. However, when we looked for a Moco-related phenotype in ATM1 and ATM2 T-DNA insertion lines of Arabidopsis (atm1-1, SALK_090939; atm1-2, SALK_121795; atm2, WiscDsLox293-296invB11), neither reduction of Mo enzyme activities and MPT/Moco levels, nor accumulation of cPMP, as are typical for ATM3-deficient plants, could be observed (see Supplemental Figure 3 online). In addition, atm3 atm1 double mutants had phenotypes identical to atm3 mutants rather than an intensification of the atm3 phenotype (Bernard et al., 2009). Hence, these observations indicate that neither ATM1 nor ATM2 participate in the transport of cPMP, and they suggest that the transfer of cPMP across the inner mitochondrial membrane of atm3 mutants must be facilitated by other mechanisms. In this respect, the possibility of cPMP being able to pass the inner membrane simply by diffusion has to be considered. In fact, cPMP has been reported to be capable of diffusing between human cells from type B patients, which have a deficiency in the conversion of cPMP to MPT, and cocultured cells from type A patients, which are impaired in cPMP synthesis (Johnson et al., 1989). Since type A cells regained SO activity after cocultivation, it was concluded that these cells absorbed cPMP (referred to as Molybdopterin precursor) synthesized by type B cells and further processed it to Moco. These experiments demonstrated not only that cPMP has the potential to diffuse across membranes but also that cPMP is stable for a time long enough to allow conversion into MPT and Moco after the diffusion process. The same properties of cPMP are also suggested by the recently reported therapy of Moco-deficient mice (Schwarz et al., 2004). Here, a mouse model was established that is characterized by a knockout of the MOCS1 gene and thus is unable to synthesize cPMP and all further downstream Moco derivatives. Newborn mice homozygous for the MOCS1 knockout died within 11 d after birth without any treatment; however, the mutation was fully rescued when purified cPMP was injected intrahepatically every other day. As shown for the reconstitution of type A cells by Johnson et al. (1989), intrahepatically applied cPMP must have passed through membranes in the MOCS1 knockout mice to allow systemic Moco biosynthesis and reconstitution of Mo-enzyme activities. Although the presence of other compensatory systems cannot be excluded, it is thus reasonable to assume from these observations that cPMP in mitochondria of atm3 mutants passively passes through the mitochondrial membranes, at least to a certain extent, to allow synthesis of Moco outside the mitochondria. In fact, when purified cPMP was added to protoplasts prepared from C24 and atm3-1 suspension cultures, a reconstitution of MPT/Moco levels was observed in the mutant, whereas no further increase in MPT/Moco amounts was detected in the C24 wild type (Figure 9). These findings show that cPMP is capable of passing biological membranes even without a functional transporter, not only in mammals, but also in plants, and they confirm that the mutation in ATM3 affects allocation of cPMP rather than its conversion into MPT.
Figure 9.
Reconstitution of Moco Biosynthesis by Coincubation of atm3-1 Protoplasts with cPMP.
Protoplasts of Arabidopsis C24 wild type and the atm3-1 mutant were coincubated with 2.5 μ g of purified cPMP for the indicated periods. Cells were harvested by centrifugation, broken by sonication, and subjected to MPT/Moco (FormA dephospho) analysis. MPT/Moco amounts were calculated to 1 mg of total protein in the protoplast extracts (n = 3; bars represent se of the means).
Besides its suggested roles in the maturation of extramitochondrial Fe-S proteins (Kushnir et al., 2001; Chen et al., 2007; Bernard et al., 2009) and heavy metal resistance (Kim et al., 2006), our study has now demonstrated that Arabidopsis ATM3 plays an important role also in Moco biosynthesis, namely, by its involvement in the transport of cPMP from mitochondria into the cytosol. If ATM3 transports cPMP directly, two different possibilities have to be considered: (1) ATM3 facilitates transport of cPMP totally independently of the transport of the Fe-S cluster precursors. This case would require ATM3 to accept different substrates, which has already been shown for other ABC transporters (summarized in Rea et al., 1998). (2) ATM3 transports an as yet unknown compound as its specific substrate that can cotransport Fe-S precursors and cPMP. The substrates of the human and yeast ATM3 homologous proteins, ABCB7 and Atm1p, respectively, which both are known to be required for the maturation of extramitochondrial Fe-S clusters (Lill and Muehlenhoff, 2008), are indeed unknown, and further work is required to identify this compound and to shed light on the substrate preference of ATM3 and its human and yeast homologs.
METHODS
Plant Material
Arabidopsis thaliana plants were grown in potting soil in a growth chamber under a constant day/night regime of 16 h light/8 h darkness at an ambient temperature of 21°C and an ambient air humidity of 50%. Leaf material was harvested after 5 weeks when cPMP accumulation was found to be most pronounced and stored at −70°C until further use.
Preparation of Arabidopsis Protoplasts
Protoplasts were isolated from fine dispersed suspensions, which were established from leaf-derived callus of the Arabidopsis C24 wild type and atm3-1 mutant. Cells were grown in liquid NH4Su medium (Grafe and Müeller, 1983) supplemented with 40 mM NH4 succinate and 20 mM KCl on a rotary shaker at 120 rpm and 23°C in darkness and subcultured once a week. For protoplast isolation, cells were incubated with gentle shaking at 50 rpm at 25°C in an enzyme solution containing 0.6% (w/v) Onozuka Cellulase R10 (Duchefa) and 0.2% (w/v) Macerocyme (Duchefa) dissolved in T0 (Crepy et al., 1982) but with Tween 80 omitted. After 14 to 16 h, isolated protoplasts were separated from undigested cell aggregates by filtration through 40- μ m sieves, washed twice with W5 solution (Negrutiu et al., 1987), and centrifuged (100g, 5 min), and aliquots of 8 × 106 protoplasts were resuspended in 5 mL of W5 for treatment with cPMP.
Treatment of Protoplasts with Purified cPMP
Isolated protoplasts of Arabidopsis wild-type C24 and the atm3-1 mutant (∼8 × 106 cells for each experiment) were coincubated with 2.5 μ g of purified cPMP prepared as described earlier (Santamaria-Araujo et al., 2004). Immediately after incubation at 22°C for 0, 30, or 60 min, respectively, cells were broken by sonication and subjected to FormA dephospho analysis as described before (Johnson and Rajagopalan, 1982; Schwarz et al., 1997).
Determination of Protein Concentrations
Concentrations of total soluble protein were determined using Roti Quant solution (Roth) according to Bradford (1976).
Chemical Detection of Moco, MPT, and cPMP
Moco and its metal-free precursors MPT and cPMP were detected by converting them to their stable oxidation products FormA-dephospho (for Moco and MPT) and CompoundZ (for cPMP) according to Johnson and Rajagopalan (1982). Oxidation, dephosphorylation, QAE chromatography, and HPLC analysis were performed basically as described by Schwarz et al. (1997). Oxidation was performed with 100 to 400 μ g plant crude extract in a total volume of 400 μ L 0.1 M Tris-HCl, pH 7.2, by the addition of 50 μ L of acidic iodine. After overnight oxidation, excess iodine was removed by the addition of 56 μ L 1% ascorbic acid, and the sample was adjusted to pH 8.3 with 1 M Tris solution. The phosphate monoester of FormA was cleaved by calf intestine alkaline phosphatase to obtain FormA-dephospho. FormA-dephospho and CompoundZ were further purified on QAE Sephadex A-25 columns. Subsequently, FormA-dephospho was eluted in a volume of 800 μ L acetic acid prior to elution of CompoundZ in 900 μ L of 50 mM HCl. The resulting fractions were analyzed by injection onto a C-18 reversed-phase HPLC column.
Enzyme Assays
For determination of XDH and AO activities, Arabidopsis leaves were squeezed at 4°C in two volumes of extraction buffer (100 mM potassium phosphate, 2.5 mM EDTA, and 5 mM DTT, pH 7.5), sonicated, and centrifuged. Enzyme activities of AO and XDH in the resulting supernantant were detected after native PAGE by activity staining with indole-3-carboxaldehyde and 1-naphthaldeyhde as substrates for AO or hypoxanthine as substrate for XDH as previously described by Koshiba et al. (1996) and Hesberg et al. (2004). For determination of NR activity, 100 mg of leaf material was ground in liquid nitrogen, and the assay was performed as described by Scheible et al. (1997). For determination of SO activity, leaf extracts were prepared as follows. One hundred milligrams of leaf material was ground in liquid nitrogen, 400 μ L 0.1 M Tris acetate, pH 7.2, was added per sample, and samples were sonicated and then centrifuged for 20 min at 21,000g and 4°C. One milliliter of saturated ammonium acetate solution was added to 400 μ L of the supernatant at 4°C. After centrifugation, the protein pellet was resuspended in 400 μ L 0.1 M Tris acetate, pH 7.2, and desalted on a Sephadex G-50 Nick gel filtration column (Amersham/Pharmacia). SO activity in these extracts was measured as described by Lang et al. (2007).
Isolation of Mitochondria
Mitochondria from green Arabidopsis leaves were isolated according to Day et al. (1985) modified by H.P. Braun (University of Hannover, Germany). Briefly, 50 to 100 g of 5-week-old plant material were homogenized at 4°C in 2 to 3 volumes of grinding buffer (0.3 M mannitol, 25 mM sodium pyrophosphate, 10 mM KH2PO4, 1 mM EDTA, 0.2% BSA, 0.5% polyvinylpyrrolidone-40, and 4 mM l-cysteine, pH 7.6). The homogenate was filtered through muslin and centrifuged at 2000g. After a second centrifugation step at 12,000g, the pellet was resuspended in 30 to 50 mL resuspension buffer (0.3 M mannitol, 10 mM KH2PO4, 2 mM glycine, and 0.1% BSA, pH 7.2). Both centrifugation steps were repeated and the pellet was homogenized in 1 to 2 mL resuspension buffer with a small douncer. The resulting homogenate was layered on a linear gradient of polyvinylpyrrolidone-20 (0 to 10%) and was ultracentrifuged at 75,000g for 45 min. The bottom end of the gradient contained the mitochondria, which were removed and washed several times with 10 volumes of resuspension buffer that did not contain BSA. The isolated mitochondria were stored at –70°C until further use. The integrity of atm3-1 mitochondria is shown in Supplemental Figure 4 online and extensively described by Bernard et al. (2009). The integrity of mitochondria prepared from the cnx5 mutant is shown in Supplemental Figure 5 online.
Isolation and Subfractionation of Mitochondria from Cauliflower
Mitochondria from cauliflower (Brassica oleracea) were isolated and subfractionated basically as described by Millar et al. (2001). Approximately 1 to 1.5 kg of cauliflower material was homogenized in 500 mL of grinding buffer (0.3 M mannitol, 50 mM sodium pyrophosphate, 0.2% BSA, 0.5% polyvinylpyrrolidone-40, 2 mM EGTA, and 20 mM l-cysteine, pH 7.5) using an electric juice extractor. The resulting homogenate was filtered through muslin and centrifuged at 1000g. The supernatant was then centrifuged at 15,000g. The pellet was resuspended in washing buffer (0.3 M mannitol and 20 mM sodium pyrophosphate, pH 7.5) and layered onto a stepped gradient of Percoll consisting of steps of 50, 28, and 20% (v/v) Percoll in 0.3 M mannitol. After centrifugation at 40,000g for 45 min, mitochondria were recovered from the 28%/50% Percoll interface. Mitochondria were washed twice in washing buffer by centrifugation at 15,000g. Mitochondria were either directly frozen at −70°C as pellets or resuspended in 0.3 M mannitol, 10 mM KH2PO4, and 2 mM glycine prior to freezing.
For subfractionation, mitochondrial pellets were resuspended in subfractionation buffer (20 mM MOPS and 70 mM sucrose, pH 7.2), the final protein concentrations were adjusted to 10 mg/mL, and the samples were incubated on ice for 20 min. After centrifugation at 15,000g for 15 min, the supernatant containing outer membrane and intermembrane space was transferred to new tubes, while the pellets, which contained mitoplasts, were resuspended in 10 mM MOPS in the same volume as before and ruptured by three rounds of freeze-thawing in liquid nitrogen. By centrifugation at 15,000g for 15 min, the inner mitochondrial membrane (pellet) was devided from the mitochondrial matrix (supernatant). All samples were stored at –70°C until further use.
Immunoblot Analysis
For immunoblot analysis, protein extracts were electrophoresed either on 12 to 15% polyacrylamide gels in the presence of SDS (for detection of CNX2, CNX3, Porin, Isu1, PetC, NBP35, AOX, Aco1, Rieske, E1a, Cyt c, SO, and NR) or on nondenaturing 7.5% polyacrylamide gels in the absence of SDS (for detection of AO and XDH). After blotting proteins onto PVDF membrane, the respective primary antibody from rabbit was used.
Antibodies Used in This Study
Polyclonal CNX2 and CNX3 antibodies were generated using recombinant CNX2 and CNX2 proteins. Cloning, expression, and purification of CNX2 and CNX3 proteins as antigen for injection into rabbits were as follows: total RNA from Arabidopsis Col-0 roots was prepared using the NucleoSpin RNA Plant Kit from Macherey-Nagel and reverse transcribed in the presence of a PolyT18 primer and MMLV reverse transcriptase (Promega). cDNAs with full-length open reading frames of CNX2 and CNX3 were obtained by RT-PCR using a 1:10 mixture of Sawady Pwo polymerase and Taq polymerase (Peqlab), forward primer CNX2_ATG_BamHI (5′-CCAAAGTGTTGGGATCCATGAGGAGGTGCTTCT-3′) and reverse primer CNX2-STOP _BamHI (5′-CCGGAGCTTAGGATCCAATGTGTATCATTGGTCTATTAGCTG-3′) for amplification of CNX2, and forward primer CNX3_ATG (5′-ATATATGGATCCATGATTTCGACGCTCCGTCGCGCCG-3′) and reverse primer CNX3_STOP (5′-ATATATGGATCCGAGTCTAGACCAAGACCCGCTCTTTCC-3′) for amplification of CNX3. The obtained RT-PCR products were visualized on 1% agarose gels in the presence of ethidium bromide and excised from the gel by use of the Nucleo Spin Extract kit (Macherey and Nagel). In a subsequent PCR reaction with Pwo polymerase and primers CNX3_ATG-BamHI (5′-TATATAGGATCCATGTCTAAGCTAACACATGTTGGTATC-3′) and CNX3_TAG_BamHI (5′-TATATAGGATCCCTAGAGTCTAGACCAAGACCCGCT-3′), the putative targeting signal was removed from the CNX3 coding sequence. The resulting PCR fragments were cloned into the BamHI restriction site of the pQE80 plasmid (Qiagen), which allows expression of 6xHis-tag fusion proteins in Escherichia coli, and correctness was confirmed by sequencing (GATC). For protein expression, cells, transformed either with pQE80/CNX2 or with pQE80/CNX3, were grown at 37°C to an OD600 of 0.7 prior to induction with 0.2 mM isopropyl- β-thiogalactoside. After induction, cells were cultured for an additional 20 h at 22°C. Cells were harvested by centrifugation and stored at –70°C until use. Purification of recombinant CNX2 and CNX3, respectively, was performed on a Ni-nitrilotriacetic acid-superflow matrix (Qiagen) under native conditions at 4°C according to the manufacturer's instructions. The purity of the resulting protein fractions was analyzed by SDS-PAGE and Coomassie staining (see Supplemental Figure 6A online). Fractions with highest purity and protein concentration were chosen for generation of CNX2 or CNX3 antibodies in rabbit (Sigma-Genosys). For immunoblot analysis, polyclonal anti-CNX2 and anti-CNX3 antibodies were diluted 1:2500 in LowCross buffer (Diagonal).
Polyclonal AAO1 antibodies were generated by Bioscience. Recombinant Arabidopsis AAO1 protein, which was used as antigen for injection into rabbits, was expressed and purified as described by Koiwai et al. (2000). AAO1 antibodies were diluted 1:1000 in standard TBST buffer containing 5% milk powder. Polyclonal antibodies raised against XDH from Lens esculenta were kindly provided by Paolo Montalbini (Perugia, Italy) and used in a 1:1000 dilution in TBST buffer containing 5% milk powder. Antibodies raised against a conserved peptide of Arabidopsis NR proteins, NIA1 and NIA2, were purchased from Antikoerper-Online and used in a 1:1000 dilution in TBST buffer containing 5% milk powder. Specificity of anti-CNX2, anti-CNX3, anti-AAO1, anti-XDH, and anti-NR antibodies is shown in Supplemental Figure 6 online. Antibodies used for detection of Arabidopsis SO were used as described in a previous study (Eilers et al., 2001). Antisera against the marker proteins NBP35, PetC, and Isu1 from Arabidopsis have been described previously (Bych et al., 2008). Antibodies against potato (Solanum tuberosum) porin have likewise been described in a previous study (Heins et al., 1994). Antibodies against the marker proteins Idh (isocitrate dehydrogenase) and cytochrome c oxidase subunit II (CoxII) were purchased from Antibody-Online. As secondary antibody, a horseradish peroxidase–conjugated anti-rabbit Ig was used (Sigma-Aldrich; 1:10,000 dilution) to detect chemiluminescence with the ECL system (Amersham Biosciences), except for the anti-AO and anti-XDH antibodies, where an alkaline phosphatase–coupled secondary antibody was used in combination with the BCIP/NBT system (Promega).
Preparation of Total RNA and Genomic DNA
Total RNA from Arabidopsis leaves was prepared using the NucleoSpin RNA plant kit (Macherey and Nagel) according to the manufacturer's instructions. Genomic DNA from Arabidopsis leaves was prepared using the innuPREP Plant DNA kit (AnalytikJena).
RT-PCR and Inverse PCR
For each reverse transcriptase reaction, 1 μ g of Arabidopsis total RNA was reverse transcribed with AMV reverse transcriptase (Promega) and oligo-d(T)18 primer according to standard procedures. Subsequent PCR was performed on a PCR Express gradient cycler using GoTaq DNA polymerase (Promega). ATM3-specific primers were atm3_start (5′-GATGTCGAGAGGATCTCGATTCG-3′) and atm3_stop (5′-CACATTTTGGATCCGAGGTTCACTATTCCAATTTGATAGC-3′). The following PCR program was used: 5 min at 94°C for initial denaturing of templates, 35 cycles including denaturing for 1 min at 94°C, annealing for 1 min at 56°C and elongation for 2 min at 72°C, and a final elongation step for 7 min at 72°C. RT-PCR generated fragments were directly ligated to pJet/blunt 1.2 vector (MBI Fermentas) and sequenced (GATC) for ensuring proper amplification.
For inverse DNA, 200 ng of genomic DNA from atm3-2 and control plants was digested with HindIII endonuclease, which does not cut within the pGABI1 T-DNA. After removing HindIII by phenole/chloroform treatment and DNA precipitation, the DNA was coincubated with T4-DNA ligase in a volume of 0.5 mL to allow circularization of single DNA fragments. After a second phenol/chloroform treatment and DNA precipitation, the DNA was resuspended in 10 μ L sterile water and was used as template for inverse PCR. The primers used to amplify the genomic DNA between right and left border of the pGABI1 T-DNA were pGABI1_LB1 (5′-ATATTGACCATCATACTCATTGC-3′) and pGABI1_RB1 (5′-CCTGTAAAGGATCTGGGTCCAGC-3′). The obtained PCR products were ligated into pJet/blunt 1.2 vector and subjected to sequence analysis (GATC), which demonstrated that atm3-2 plants carry a single T-DNA insertion in the ATM3 locus.
ABA Determination
Arabidopsis leaves were ground in liquid nitrogen, homogenized in distilled water at a ratio of 1:7 (w/v), and placed overnight in the dark at 4°C. The extracts were centrifuged at 10,000g for 10 min at 4°C, and the resulting supernatant was diluted four times in standard TBS buffer. ABA in these extracts was quantified using the Phytodetek ABA ELISA kit (Agdia/Linaris) according to the manufacturer's instructions.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers NP_850177 (CNX2, AGI locus AT2G31955), NP_171636 (CNX3, AGI locus AT1G01290), and NP_200635 (STA1/ATM3, AGI locus AT5G58270).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Activities and Immunoanalysis of Molybdoproteins in Moco-Deficient Mutants.
Supplemental Figure 2. Relative Expression of CNX Genes in Leaves of the C24 Wild Type and the atm3-1 Mutant.
Supplemental Figure 3. Biochemical Characterization of the Arabidopsis Insertion Lines atm1-1, atm1-2, and atm2.
Supplemental Integrity of Mitochondria from C24 Wild Type and the atm3-1 Mutant.
Supplemental Figure 5. Integrity of Mitochondria from Col-0 Wild Type and the sir1/cnx5 Mutant.
Supplemental Figure 6. Specificity of Antibodies Used in This Study.
Supplemental References.
Acknowledgments
We thank Sergei Kushnir (University of Gent, Belgium) for the sta1 mutant line, Yunde Zhao (University of California at San Diego/La Jolla, CA) for providing sir1/cnx5, sir5/cnx7, and sir/cnx1 mutant seeds, and Paolo Montalbini (University of Perugia, Italy) for providing XDH antiserum. We also thank Hans-Peter Braun (University of Hannover, Germany) for kindly providing antiporin antibodies and Victoria Michael, Marion Kay, Ute Nielaender, and Anne Reichelt (Technische Universität Braunschweig) for excellent technical assistance. This work was funded by a grant from the Deutsche Forschungsgemeinschaft to R.R.M. and F.B.
References
- Akaba S., Seo M., Dohmae N., Takio K., Sekimoto H., Kamiya Y., Furuya N., Komano T., Koshiba T. (1999). Production of homo- and hetero-dimeric isozymes from two aldehyde oxidase genes of Arabidopsis thaliana. J. Biochem. 126: 395–401 [DOI] [PubMed] [Google Scholar]
- Allikmets R., Raskind W.H., Hutchinson A., Schueck N.D., Dean M., Koeller D.M. (1999). Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8: 743–749 [DOI] [PubMed] [Google Scholar]
- Babiychuk E., Fuangthong M., Van Montagu M., Inzé D., Kushnir S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94: 12722–12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Lobréaux S. (2005). Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 10: 324–331 [DOI] [PubMed] [Google Scholar]
- Bernard D.G., Cheng Y., Zhao Y., Balk J. (2009). An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol. 151: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bych K., Netz D.J., Vigani G., Bill E., Lill R., Pierik A.J., Balk J. (2008). The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 283: 35797–35804 [DOI] [PubMed] [Google Scholar]
- Chen S., Sánchez-Fernández R., Lyver E.R., Dancis A., Rea P.A. (2007). Functional characterization of AtATM1, AtATM2, and AtATM3, a subfamily of Arabidopsis half-molecule ATP-binding cassette transporters implicated in iron homeostasis. J. Biol. Chem. 282: 21561–21571 [DOI] [PubMed] [Google Scholar]
- Crawford N.M. (1992). Study of chlorate-resistant mutants of Arabidopsis: Insights into nitrate assimilation and ion metabolism of plants. Genet. Eng. (N. Y.) 14: 89–98 [Google Scholar]
- Crepy L., Chupeau M.-C., Chupeau Y. (1982). The isolation and culture of leaf protoplasts of Cichorium intybus and their regeneration into plants. Z. Pflanzenphysiol. 107: 123–131 [Google Scholar]
- Dai X., Hayashi K., Nozaki H., Cheng Y., Zhao Y. (2005). Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day D.A., Neuburger M., Douce R. (1985). Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust. J. Plant Physiol. 12: 219–228 [Google Scholar]
- Dos Santos P.C., Dean D.R., Hu Y., Ribbe M.W. (2004). Formation and insertion of the nitrogenase iron-molybdenum cofactor. Chem. Rev. 104: 1159–1173 [DOI] [PubMed] [Google Scholar]
- Eilers T., Schwarz G., Brinkmann H., Witt C., Richter T., Nieder J., Koch B., Hille R., Haensch R., Mendel R.R. (2001). Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J. Biol. Chem. 276: 46989–46994 [DOI] [PubMed] [Google Scholar]
- Frazzon A.P., Ramirez M.V., Warek U., Balk J., Frazzon J., Dean D.R., Winkel B.S. (2007). Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol. Biol. 64: 225–240 [DOI] [PubMed] [Google Scholar]
- Garattini E., Fratelli M., Terao M. (2008). Mammalian aldehyde oxidases: Genetics, evolution and biochemistry. Cell. Mol. Life Sci. 65: 1019–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.R., Hänsch R. (2009). New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Grafe R., Müller A. (1983). Complementation analysis of nitrate reductase deficient mutants of Nicotiana tabacum by somatic hybridization. Theor. Appl. Genet. 66: 127–130 [DOI] [PubMed] [Google Scholar]
- Haenzelmann P., Hernandez H.L., Menzel C., Garcia-Serres R., Huynh B.H., Johnson M.K., Mendel R.R., Schindelin H. (2004). Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 279: 34721–34732 [DOI] [PubMed] [Google Scholar]
- Haenzelmann P., Schindelin H. (2004). Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl. Acad. Sci. USA 101: 12870–12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenzelmann P., Schwarz G., Mendel R.R. (2002). Functionality of alternative splice forms of the first enzymes involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 277: 18303–18312 [DOI] [PubMed] [Google Scholar]
- Havemeyer A., Bittner F., Wollers S., Mendel R.R., Kunze T., Clement B. (2006). Identification of the missing component in the mitochondrial benzamidoxime prodrug converting system as a novel molybdenum enzyme. J. Biol. Chem. 281: 34796–34802 [DOI] [PubMed] [Google Scholar]
- Heins L., Mentzel H., Schmid A., Benz R., Schmitz U.K. (1994). Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J. Biol. Chem. 269: 26402–26410 [PubMed] [Google Scholar]
- Hesberg C., Haensch R., Mendel R.R., Bittner F. (2004). Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: Differential gene expression and enzyme activities. J. Biol. Chem. 279: 13547–13554 [DOI] [PubMed] [Google Scholar]
- Hoff T., Schnorr K.M., Meyer C., Caboche M. (1995). Isolation of two Arabidopsis cDNAs involved in early steps of molybdenum cofactor biosynthesis by functional complementation of Escherichia coli mutants. J. Biol. Chem. 270: 6100–6107 [DOI] [PubMed] [Google Scholar]
- Ibdah M., Chen Y.T., Wilkerson C.G., Pichersky E. (2009). An aldehyde oxidase in developing seeds of Arabidopsis converts benzaldehyde to benzoic acid. Plant Physiol. 150: 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Rajagopalan K.V. (1982). Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc. Natl. Acad. Sci. USA 79: 6856–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Wuebbens M.M., Mandell R., Shih V.E. (1989). Molybdenum cofactor biosynthesis in humans. Identification of two complementation groups of cofactor-deficient patients and preliminary characterization of a diffusible molybdopterin precursor. J. Clin. Invest. 83: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Duran M. (2001). Molybdenum cofactor deficiency and isolated sulfite oxidase deficiency. The Metabolic and Molecular Bases of Inherited Disease, Scriver C., Beaudet A., Sly W., Valle D., (New York: McGraw-Hill; ), pp. 3163–3177 [Google Scholar]
- Kim D.Y., Bovet L., Kushnir S., Noh E.W., Martinoia E., Lee Y. (2006). AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 140: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Csere P., Prohl C., Lill R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai H., Akaba S., Seo M., Komano T., Koshiba T. (2000). Functional expression of two Arabidopsis aldehyde oxidases in the yeast Pichia pastoris. J. Biochem. 127: 659–664 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Saito E., Ono N., Yamamoto N., Sato M. (1996). Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptiles of maize. Plant Physiol. 110: 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper J., Llamas A., Hecht H.J., Mendel R.R., Schwarz G. (2004). Structure of molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430: 803–806 [DOI] [PubMed] [Google Scholar]
- Kushnir S., Babiychuk E., Storozhenko S., Davey M.W., Papenbrock J., De Rycke R., Engler G., Stephan U.W., Lange H., Kispal G., Lill R., Van Montagu M. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C., Popko J., Wirtz M., Hell R., Herschbach C., Kreuzwieser J., Rennenberg H., Mendel R.R., Haensch R. (2007). Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ. 30: 447–455 [DOI] [PubMed] [Google Scholar]
- Lardi-Studler B., Smolinsky B., Petitjean C.M., Koenig F., Sidler C., Meier J.C., Fritschy J.M., Schwarz G. (2007). Vertebrate-specific sequences in the gephyrin E-domain regulate cytosolic aggregation and postsynaptic clustering. J. Cell Sci. 120: 1371–1382 [DOI] [PubMed] [Google Scholar]
- Léon S., Touraine B., Briat J.F., Lobréaux S. (2005). Mitochondrial localization of Arabidopsis thaliana Isu Fe-S scaffold proteins. FEBS Lett. 579: 1930–1934 [DOI] [PubMed] [Google Scholar]
- Lill R., Muehlenhoff U. (2008). Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77: 669–700 [DOI] [PubMed] [Google Scholar]
- Llamas A., Otte T., Multhaupt G., Mendel R.R., Schwarz G. (2006). The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin: A novel route toward metal cofactor assembly. J. Biol. Chem. 281: 18343–18350 [DOI] [PubMed] [Google Scholar]
- Matthies A., Nimtz M., Leimkuehler S. (2005). Molybdenum cofactor biosynthesis in humans: Identification of a persulfide group in the rhodanese-like domain of MOCS3 by mass spectrometry. Biochemistry 44: 7912–7920 [DOI] [PubMed] [Google Scholar]
- Matthies A., Rajagopalan K.V., Mendel R.R., Leimkuehler S. (2004). Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc. Natl. Acad. Sci. USA 101: 5946–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhorn V., Matsumi K., Koiwai H., Ikegami K., Okamoto M., Nambara E., Bittner F., Koshiba T. (2008). Transient expression of AtNCED3 and AAO3 genes in guard cells causes stomatal closure in Vicia faba. J. Plant Res. 121: 125–131 [DOI] [PubMed] [Google Scholar]
- Mendel R.R., Bittner F. (2006). Cell biology of molybdenum. Biochim. Biophys. Acta 1763: 621–635 [DOI] [PubMed] [Google Scholar]
- Millar A.H., Liddell A., Leaver C.J. (2001). Isolation and subfractionation of mitochondria from plants. Methods Cell Biol. 65: 53–74 [DOI] [PubMed] [Google Scholar]
- Negrutiu I., Shillito R., Potrykus I., Biasini G., Sala F. (1987). Hybrid genes in the analysis of transformation conditions. I. Setting up a simple method for direct gene transfer in plant protoplasts. Plant Mol. Biol. 8: 363–373 [DOI] [PubMed] [Google Scholar]
- Pilon M., Abdel-Ghany S.E., Van Hoewyk D., Ye H., Pilon-Smits E.A. (2006). Biogenesis of iron-sulfur cluster proteins in plastids. Genet. Eng. (N. Y.) 27: 101–117 [DOI] [PubMed] [Google Scholar]
- Rea P.A., Li Z.S., Lu Y.P., Drozdowicz Y.M., Martinoia E. (1998). From vacuolar GS-X pumps to multispecific ABC transporters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 727–776 [DOI] [PubMed] [Google Scholar]
- Santamaria-Araujo J.A., Fischer B., Otte T., Nimtz M., Mendel R.R., Wray V., Schwarz G. (2004). The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J. Biol. Chem. 279: 15994–15999 [DOI] [PubMed] [Google Scholar]
- Scheible W.R., Lauerer M., Schulze E.D., Caboche M., Stitt M. (1997). Accumulation of nitrate in the shoot acts as a signal to regulate shoot:root allocation in tobacco. Plant J. 11: 671–691 [Google Scholar]
- Schwarz G. (2005). Molybdenum cofactor biosynthesis and deficiency. Cell. Mol. Life Sci. 62: 2792–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G., Boxer D.H., Mendel R.R. (1997). Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 272: 26811–26814 [DOI] [PubMed] [Google Scholar]
- Schwarz G., et al. (2004). Rescue of lethal molybdenum cofactor deficiency by a biosynthetic precursor from Escherichia coli. Hum. Mol. Genet. 13: 1249–1255 [DOI] [PubMed] [Google Scholar]
- Seo M., Koiwai H., Akaba S., Komano T., Oritani T., Kamiya Y., Koshiba T. (2000a). Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 23: 481–488 [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). The complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Seo M., Peeters A.J., Koiwai H., Oritani T., Marion-Poll A., Zeevaart J.A., Koornneef M., Kamiya Y., Koshiba T. (2000b). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier P.J., et al. (2008). Plant ABC proteins: A unified nomenclature and updated inventory. Trends Plant Sci. 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Zhu J.K. (2005). Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem. Soc. Trans. 33: 375–379 [DOI] [PubMed] [Google Scholar]
- Wuebbens M.M., Liu M.T., Rajagopalan K.V., Schindelin H. (2000). Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC. Structure 8: 709–718 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Dai X., Blackwell H.E., Schreiber S.L., Chory J. (2003). SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]