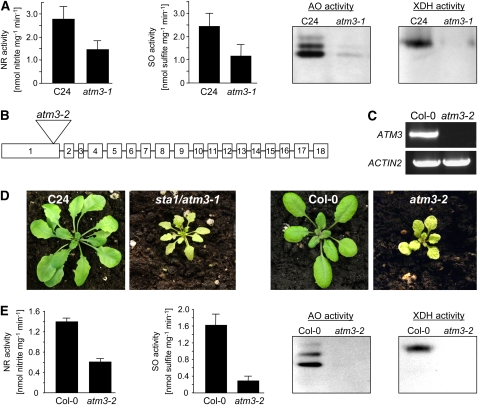
Figure 1.
Phenotypical Characterization of atm3-1 and atm3-2 Mutants.
(A) Mo enzyme activities (NR, SO, AO, and XDH) in leaves of the sta1/atm3-1 mutant and the C24 wild type (NR, n = 4; SO, n = 6; bars represent se of the means). For comparison of AO and XDH activities, leaf extracts were electrophoresed on native polyacrylamide gels and either stained for AO activity or for XDH activity. For each, one representative gel of six experiments is shown.
(B) T-DNA insertion site in the Arabidopsis GABI-Kat atm3-2 mutant.
(C) RT-PCR–based detection of full-length ATM3 trancripts in Columbia-0 (Col-0) wild types and atm3-2 mutant plants (n = 3). Actin2 primers were used to demonstrate successful reverse transcription of mRNA.
(D) Phenotypical comparison of atm3-1 and atm3-2 mutants.
(E) Mo enzyme activities (NR, SO, AO, and XDH) in leaves of the GABI-Kat atm3-2 mutant and the Col-0 wild type (NR, n = 4; SO, n = 3; bars represent seof the means). For AO and XDH activities, one representative gel of four experiments is shown.