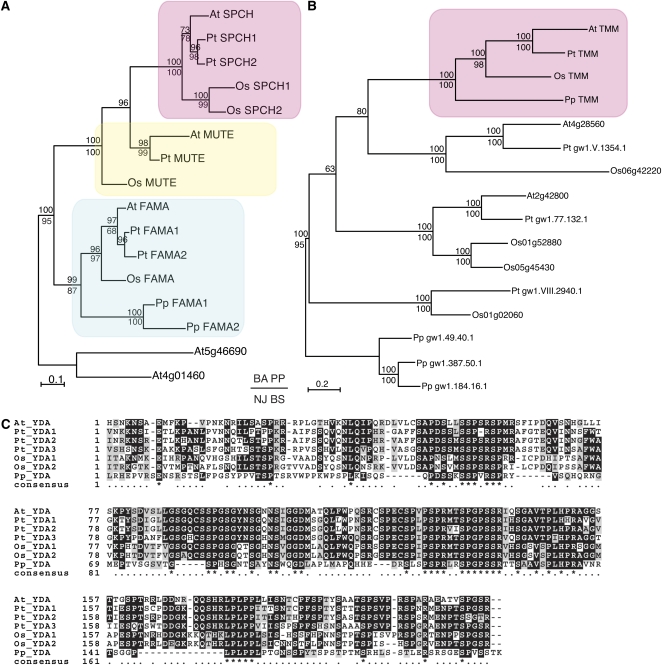
Figure 6.
Stomatal Patterning and Spacing Genes Are Conserved among Embryophytes.
(A) A gene tree of master regulatory bHLH transcription factors (SPCH, MUTE, and FAMA) in stomatal development. Amino acid sequences from bHLH and ACT domains were aligned and the tree constructed using Bayesian and neighbor-joining methods. The Bayesian phylogram is shown. The pink, yellow, and cyan rectangles highlight the SPCH, MUTE, and FAMA clades, respectively. Posterior probability values are indicated above the nodes, and bootstrap values over 50 (100,000 replicates) are indicated below the nodes.
(B) A gene tree of TMM, which encodes an LRR receptor-like protein necessary for proper stomatal spacing in Arabidopsis. The entire amino acid sequence was aligned and the tree constructed as in (A). The Bayesian phylogram is shown. The pink rectangle highlights the single copy TMM clade. Posterior probability values are indicated above the nodes, and bootstrap values over 50 (100,000 replicates) are indicated below the nodes.
(C) Amino acid alignment of the N-terminal portion of YODA (YDA), a MAPKKK, which is essential to regulate the activity of the protein in Arabidopsis. YDA is also required for appropriate stomatal spacing. At, Arabidopsis thaliana; Pt, Populus trichocarpa; Os, Oryza sativa; Pp, Physcomitrella patens. See Supplemental Methods and Supplemental Table 1 online for detailed bioinformatic and phylogenetic methods and gene ID numbers.