Abstract
The peroxisome proliferator-activated receptor γ coactivator-1 (PGC-1) family is a key regulator of mitochondrial function, and reduced mRNA expression may contribute to muscle lipid accumulation in obesity and type 2 diabetes. To characterize the effects of PGC-1 on lipid metabolism, we overexpressed PGC-1α and PGC-1β in C2C12 myotubes using adenoviral vectors. Both PGC-1α and -1β increased palmitate oxidation [31% (P<0.01) and 26% (P<0.05), respectively] despite reductions in cellular uptake [by 6% (P<0.05) and 21% (P<0.001)]. Moreover, PGC-1α and -1β increased mRNA expression of genes regulating both lipid oxidation (e.g., CPT1b and ACADL/M) and synthesis (FAS, CS, ACC1/2, and DGAT1). To determine the net effect, we assessed lipid composition in PGC-1-expressing cells. Total lipid content decreased by 42% in palmitate-loaded serum-starved cells overexpressing PGC-1α (P<0.05). In contrast, in serum-replete cells, total lipid content was not significantly altered, but fatty acids C14:0, C16:0, C18:0, and C18:1 were increased 2- to 4-fold for PGC-1α/β (P<0.05). Stable isotope-based dynamic metabolic profiling in serum-replete cells labeled with 13C substrates revealed both increased de novo fatty acid synthesis from glucose and increased fatty acid synthesis by chain elongation with either PGC-1α or -1β expression. These results indicate that PGC-1 can promote both lipid oxidation and synthesis, with net balance determined by the nutrient/hormonal environment.—Espinoza, D. O., Boros, L. G., Crunkhorn, S., Gami, H., Patti, M.-E. Dual Modulation of both lipid oxidation and synthesis by peroxisome proliferator-activated receptor-γ coactivator-1α and -1β in cultured myotubes.
Keywords: metabolomics, fatty acid metabolism, lipid metabolism
The peroxisomal proliferator-activated receptor coactivator 1 (PGC-1) family of transcriptional coactivators, including PGC-1α, PGC-1β, and PGC-1-related coactivator (official gene names: PPARGC1A, PPARGC1B, and PPRC1), plays a central role in regulating transcription of genes critical for mitochondrial oxidative metabolism. These effects may be mediated by coactivation of key transcription factors, including PPARα (1), hepatic nuclear factor 4 (2), nuclear respiratory factors (NRFs) (3), estrogen receptor-related receptor (ERR)-α (official name ESRRA) (4), sterol regulatory element-binding protein 1 (5), myocyte enhancing factor 2 (6, 7), and forkhead box O1 (8). Moreover, PGC-1α and PGC-1β coactivate their own promoters, in part through ERRα, in a positive feedback loop (4, 7, 9, 10).
The net effect of PGC-1 activity in skeletal muscle is to increase mitochondrial biogenesis and oxidative phosphorylation (11), promote fiber type switching to more oxidative type I, IIA (6), or IIX fibers (7), and increase glucose uptake and glycogen storage (12), mediated in part via increased expression of the glucose transporter GLUT4 (13). Surprisingly, the effects of transgenic expression of PGC-1α or -1β in muscle are quite variable, potentially related to differences in magnitude of expression. Modest PGC-1α overexpression in muscle in vivo increases insulin sensitivity (14). Similarly, both whole-body and muscle-specific PGC-1β transgenic mice have improved insulin sensitivity (7, 10). Conversely, PGC-1α-null mice have 30–60% reductions in muscle oxidative phosphorylation (OXPHOS) gene expression and a modest reduction in tissue ATP (15), yet are more insulin-sensitive, perhaps related to centrally mediated hyperactivity (16, 17). Mice with muscle-specific PGC-1α deletion also have impaired muscle strength and increased insulin sensitivity (18) but exhibit modest glucose intolerance owing to coexisting reductions in insulin secretion (19). Phenotypes in PGC-1β-null mice are similar but milder and typically are accompanied by normal insulin sensitivity (20,21,22).
PGC-1 has also been implicated in regulation of metabolic homeostasis in humans. For example, mRNA expression of PGC-1α and -1β is reduced by 36 and 46%, respectively, in skeletal muscle of Mexican-American individuals with a family history of type 2 diabetes mellitus (DM) or established DM (23) and by ∼20% during hyperinsulinemia in Swedish Caucasians (24). In both studies, decreased PGC-1 mRNA expression paralleled a 25–30% reduction in mRNA expression of PGC- and NRF-dependent mitochondrial OXPHOS genes. Similar reductions in both PGC-1α and -1β mRNA expression have been observed in muscle from insulin-resistant or obese normoglycemic individuals (23, 25) and in white adipose tissue of insulin-resistant (26) and morbidly obese (27) subjects. Moreover, PGC-1 expression and function are reduced in settings of lipid excess, including both genetic and dietary obesity in rodents (28, 29) and after acute high-fat feeding (30) or lipid infusion in healthy humans (31).
Although these data support the hypothesis that decreased PGC-1 expression could contribute to impaired mitochondrial oxidative gene expression/function and increased lipid accumulation characteristic of human insulin resistance (32, 33), it remains unclear whether reduced PGC-1 mRNA expression in these settings is due to lipid accumulation or coexisting insulin resistance or could play a primary role in muscle lipid metabolism. Thus, to address this question in the absence of complex systemic physiology, we expressed PGC-1α and -1β using adenoviral vectors and assessed cellular lipid metabolism, using both traditional and metabolomic approaches, in murine C2C12 myotubes.
MATERIALS AND METHODS
Reagents
[1-14C]Palmitate and d-[14C(U)]glucose were purchased from Perkin-Elmer Life and Analytical Sciences (Wellesley, MA, USA). 2-Deoxy-d-[2,6-3H]glucose was from GE Healthcare Biosciences (Piscataway, NJ, USA). [1,2-13C2]Glucose was from Cambridge Isotope Laboratories, Inc. (Cambridge, MA, USA). [13C]18-stearic acid (18:0) was from Spectra Stable Isotopes (Columbia, MD, USA), fatty acids were from Alltech Associates (Deerfield, IL, USA), Fatty acid-free BSA was from Millipore (Kankakee, IL, USA), and culture reagents were from Invitrogen (Carlsbad, CA, USA). All other chemicals were from Sigma-Aldrich Corp. (St. Louis, MO, USA). Etomoxir was obtained from Dr. H. P. O. Wolf (Byk Gulden, Konstanz, Germany).
Antibodies
Primary antibodies included anti-PGC-1α (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-FLAG (Stratagene, La Jolla, CA, USA), anti-AMP-activated protein kinase (AMPK) α, anti-phospho-AMPKα (Thr-172) (Cell Signaling Technology, Danvers, MA, USA), anti-acetyl-CoA carboxylase (ACC) and anti-phospho-ACC (Ser-79; Upstate, Charlottesville, VA, USA), and mitochondrial protein antibodies (Invitrogen). Anti-mouse IgG and anti-rabbit IgG (horseradish peroxidase-conjugated) secondary antibodies were from New England Biolabs (Ipswich, MA, USA).
Myotube culture
Mouse C2C12 myoblasts (American Type Culture Collection, Manassas, VA, USA) were maintained subconfluent (≤70–80%) in DMEM with 25 mM glucose (DMEM-H) and 20% FBS. Differentiation medium containing 2% heat-inactivated horse serum (HS) was substituted at confluence, replaced every 2 d, and always added on the day before the experiment.
Adenoviral vectors and infection of C2C12 myotubes
Adenoviral vectors for murine green fluorescent protein (GFP), GFP-PGC-1α, and GFP-PGC-1β (9, 34) were kindly provided by Dr. Bruce Spiegelman (Dana Farber Cancer Institute, Boston, MA, USA). The PGC-1β expression vector incorporates a 3′-FLAG. Viruses were amplified in Ad-293 cells (Stratagene). Confluent myotubes were infected at differentiation days 2–3 with 1.0 × 106 PFU/cm2 of flask surface.
Small interfering RNA (siRNA)
C2C12 myotubes were transfected on d 3 with 100 nM PGC-1α/β siRNA (SMARTpools) or nontargeting siRNA (siCONTROL), using DharmaFECT 3 reagent (Dharmacon, Lafayette, CO, USA). Efficiency was determined using siGLO-RISC free nontargeting siRNA.
Palmitate and glucose oxidation
At 72 h postinfection, C2C12 myotubes were incubated with 3 ml of 1% fatty acid-free BSA-Krebs-Ringer Hepes (KRH) buffer (20 mM HEPES, 1 mM CaCl2, 1.2 mM MgSO4, 1.2 mM KH2PO4, 1.4 mM KCl, 24 mM NaHCO3, and 130 mM NaCl, pH 7.4) containing 200 nCi of either [1-14C]palmitic acid (specific activity 305 mCi/mmol) or d-[14C(U)]glucose (specific activity 50 mCi/mmol). (Please note that each isotope was used in independent experiments.) Flasks (25 cm2) were sealed with a rubber stopper with a center well (Kimble/Kontes, Vineland, NJ, USA) containing 200 μl of the CO2-trapping benzethonium. After incubation for 4 h, 300 μl of HCl was injected into the flask, the solution was equilibrated for 16 h, and relative lipid/carbohydrate oxidation was calculated from 14CO2 in center wells (Beckman LS6500; Beckman Coulter, Fullerton, CA, USA).
Glucose uptake
Differentiated myotubes were serum starved at 72 h postinfection with DMEM-H/0.1% HS for 6 or 24 h, washed twice with PBS, and incubated with 1 ml of 0.1% BSA-KRH (20 mM HEPES, pH 7.4; 136 mM NaCl; 4.7 mM KCl; 1.25 mM MgSO4; and 1.25 mM CaCl2) at 37°C for 45 min and then with 100 nM insulin or PBS for 20 min. Cytochalasin B (10 μM final) was added to control wells 5 min before insulin, and 100 μl of 10× labeling solution (1 mM 2-deoxyglucose and 5 μCi/ml of [3H[2-deoxyglucose) was added for 4 min at 37°C. Cells were washed twice with cold 10 mM glucose-PBS and lysed with 1 ml of 0.1% SDS, and 3H was measured.
Lactate secretion
Myotubes were incubated at 72 h postinfection with serum-, glucose-, and pyruvate-free DMEM for 6 h and then with DMEM-H ± 100 nM insulin. Conditioned medium was harvested at 24 h and centrifuged (16,000 g, 10 min, 4°C) for lactate measurement (Sigma-Aldrich Corp.).
Fatty acid uptake and lipid synthesis
At 48 h postinfection, myotubes were serum starved for 24 h. For fatty acid uptake assays, cells were incubated for 30 min in 1% BSA-KRH ± 100 μM etomoxir, incubated with 250 nCi/ml of [14C]palmitate for 3 min at 37°C, rinsed with 1 mM phloretin (Sigma-Aldrich Corp.)-PBS, and lysed with 0.1% SDS. Lipids were extracted with chloroform-methanol (2:1), and 14C counts were determined in the organic phase.
For lipid synthesis assays, cells were incubated with 1% BSA-KRH ± 100 nM insulin at 37°C for 15 min, and 250 nCi of [14C]glucose was added at 37°C for 1 h. Cells were washed 3 times with cold 10 mM glucose-PBS and lysed. Lipids were extracted with chloroform-methanol (2:1), and 14C counts were measured.
Real-time PCR
Total RNA was extracted using TRIzol (Invitrogen) and purified with RNeasy (Qiagen, Valencia, CA, USA). cDNA was synthesized from 1 μg of total RNA using random hexamers (Clontech, Mountain View, CA, USA) in a 100-μl reaction. PCR was performed (ABI 7000 system; Applied Biosystems, Foster City, CA, USA) using 2.5 μl of cDNA with SYBR Green: 1 cycle at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. mRNA expression was calculated relative to TATA-binding protein (TBP). Primer sequences are provided in Supplemental Table 1.
Western blotting
Proteins were extracted (35), and lysates were centrifuged at 4°C for 30 min at 16,000 g. Equal protein was separated by SDS-PAGE and transferred to 0.45-μm nitrocellulose (Bio-Rad Laboratories, Hercules, CA, USA) for subsequent Western blotting and chemiluminescence detection. ImageQuant (version 5.1; GE Healthcare Biosciences) was used for quantification.
Lipid profiling
Myotubes were incubated at 48 h postinfection with 500 μM BSA-conjugated palmitate or oleate for 24 h. Cells were rinsed and harvested into cold PBS, and pellets were frozen at −80°C. Lipid profiling was performed at the Lipid Core, Mouse Metabolic Phenotyping Center, Vanderbilt University (Nashville, TN, USA). After extraction with chloroform-methanol (2:1), individual lipid classes (phospholipids, cholesterol esters, diglycerides, and triglycerides) were separated by thin-layer chromatography, visualized by iodine vapors or rhodamine 6G, scraped, and eluted from the silica gel. Phospholipids were analyzed either in total lipid or eluted phospholipid fraction. Cholesterol esters and cholesterol were analyzed as described (36) or by gas chromatography. Lipid classes containing fatty acids were quantitated by gas chromatography (GC)/mass spectrometry (MS).
Stable isotope-based dynamic metabolic profiling (SIDMAP) and GC/MS
Myotubes were incubated at 24 h postinfection with 5 mM glucose-5 mM d-[1,2-13C2]glucose (50% enrichment) in glucose-free DMEM or 1% BSA-125 μM [13C]stearic acid in DMEM-H, with 2% HS for 72 h. Medium was replaced at 24 and 48 h with fresh labeled medium. Cells were rinsed twice and harvested into cold PBS, and pellets were frozen at −80°C. [13C]Glucose or [13C]stearate metabolites were analyzed by GC/MS. TRIzol-extracted pellets were washed with 2 ml of hexane-ethyl acetate (85:15) to elute neutral lipids. Chloroform-methanol (23:1, 4 ml) was used to extract free ceramides. Diisopropyl ether-acetic acid (98:5), acetone-methanol (9:1.35), and chloroform-methanol (2:1) were used to extract free fatty acids (FFAs), neutral glycosphingolipids and sphingoid bases, and sphingomyelin, respectively (37).
Palmitate, stearate, and long-chain fatty acids within ceramide and sphingomyelin fractions were extracted after saponification in 30% KOH and 100% ethanol using petroleum ether. Fatty acids were converted to their methylated derivative using 0.5 N methanolic HCl. Enrichment of 13C-labeled acetyl units within palmitate, stearate, and oleate were monitored at m/z 270, m/z 298, and m/z 264, respectively. Mass isotopomer distribution analysis was used to determine synthesis, elongation, and desaturation of new fatty acids within these groups. Mass spectral data were obtained on the HP5973 mass selective detector connected to an HP6890 gas chromatograph (GC inlet 250°C, transfer line 280°C, MS source 230°C, MS Quad 150°C). A Supelco (SP-2330) 30-m silica-coated column was used to separate individual fatty acids before mass isotopomer analysis.
Statistical analysis
Analysis of variance was performed using StatView 5.0.1 (SAS, Cary, NC, USA) followed by Fisher’s protected least significant difference test. For gene expression data, q values were calculated using the method of Storey (38). All values presented in figures and tables are means ± se. Some data are expressed as relative units or fold change relative to the mean of GFP-infected cells (assigned a mean value of 1), as indicated in legends for specific figures.
RESULTS
PGC-1 overexpression increases glucose oxidation in C2C12 myotubes
To determine whether experimental alterations in PGC-1 expression would modify oxidative capacity, we expressed PGC-1 using adenoviral infection in C2C12 myotubes. PGC-1α mRNA expression increased by 86-fold (P<0.0001) and that of PGC-1β mRNA by 29-fold (P<0.0001), compared with GFP controls (Supplemental Fig. 1A). These levels are 13- and 18-fold higher, respectively, than endogenous levels in fed mouse quadriceps muscle (P<0.0001 for both) (Supplemental Fig. 1A). At the protein level, PGC-1α increased by 8.5-fold compared with GFP-infected cells (P<0.01) (Supplemental Fig. 1B). Because of a lack of sensitive anti-PGC-1β antibody, recombinant PGC-1β protein was detected with an anti-FLAG antibody (not shown); thus, comparison with endogenous protein was not performed.
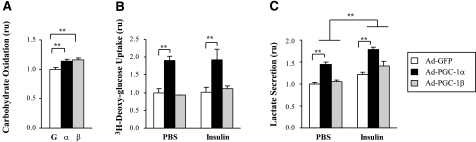
We next studied the effects of PGC-1 expression on glucose oxidation, measured by 14CO2 release after incubation with [14C(U)]glucose. Glucose oxidation increased by 14 ± 4% (P<0.01) and 15 ± 3% (P<0.01) with PGC-1α and -1β overexpression (Fig. 1A). This modest increase in glucose oxidation was probably related in part to increased glucose uptake, with a 90% increase in both basal and insulin-stimulated glucose uptake observed in cells overexpressing PGC-1α (P<0.001) (Fig. 1B) as described previously (13); no changes were observed in PGC-1β-expressing cells. [Note that insulin did not increase glucose uptake under these conditions (6 h of serum starvation) as expected, given the limited insulin-stimulated glucose uptake in C2C12 myotubes (34).] In contrast, after overnight serum starvation, insulin responsiveness was enhanced in GFP control cells (50% increase, P<0.05), but there was no significant change in basal or insulin-stimulated glucose uptake in cells overexpressing either PGC-1α or -1β (not shown). Similarly, we found no significant differences in either basal or insulin-stimulated glycogen content with either PGC-1α or -1β expression in the serum-starved state (not shown). Note that serum starvation did not alter expression of endogenous PGC-1α/β.
Figure 1.
PGC-1 overexpression effects on carbohydrate metabolism in C2C12 myotubes. A) Carbohydrate oxidation: 14CO2 release from cells was measured after incubation with d-[14C(U)]glucose. Values are means of 4 independent experiments (2 to 4 replicates each) and are presented as relative to GFP-infected cells (mean glucose oxidation 9.31±3.5 fmol [14C]glucose/μg protein/4 h. B) Glucose uptake: cells were serum starved for 6 h and then treated with 100 nM insulin or PBS for 20 min, followed by [3H]deoxyglucose for 4 min. Values are relative to GFP-infected cells. C) Lactate secretion: lactate was measured in 24-h conditioned medium. Values are means of 3 independent experiments with 3 to 5 replicas each and are relative to GFP-infected cells (mean lactate 0.1±0.06 μmol lactate/μg protein/h). Graphed data are mean ± se relative units (ru) or fold change relative to GFP-infected cells (mean value of 1). *P < 0.05; **P < 0.01.
Lactate production reflects both glycolytic oxidative metabolism and the cellular redox state. PGC-1α, but not PGC-1β, overexpression increased medium lactate in both basal and insulin-stimulated conditions [42±10% (P<0.01) and 46±21% (P<0.05), respectively] (Fig. 1C). In addition, lactate was significantly increased by insulin in PGC-1α- and -1β-infected cells (P<0.05 and P<0.01, respectively) (Fig. 1C).
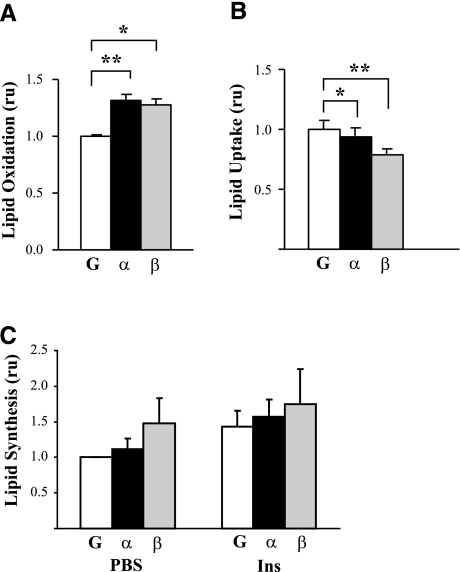
PGC-1α and PGC-1β overexpression increases palmitate oxidation in C2C12 myotubes
Because we hypothesized that alterations in PGC-1 expression and/or its regulation may be a major contributor to lipid accumulation, we tested the effect of PGC-1 expression on lipid metabolism in myotubes. Fatty acid oxidation, measured by 14CO2 release after incubation with [1-14C]palmitic acid, increased by 31 ± 7% in cells expressing PGC-1α (P<0.01) and by 26 ± 12% in cells expressing PGC-1β (P<0.05) vs. the GFP control (Fig. 2A). PGC-1-mediated increases in palmitate oxidation did not appear to be due to increased uptake from the medium, because uptake of [14C]palmitate was modestly decreased, by 6% in PGC-1α-overexpressing cells (P<0.05) and by 21% in PGC-1β-overexpressing cells (P<0.001) (Fig. 2B).
Figure 2.
PGC-1 overexpression effects on lipid oxidation and uptake. A) Lipid oxidation was measured by 14CO2 release after incubation with [14C]palmitate. Palmitate oxidation was increased by 32 ± 4% with PGC-1α (P<0.001) and by 28 ± 6% with PGC-1β (P<0.001). Values are means of 6 independent experiments with 2 to 4 replicas each and are relative to GFP-infected cells (mean palmitate oxidation 0.23 nmol/mg protein/4 h). ru, relative units. B) Lipid uptake was measured in cells serum starved for 24 h and incubated with [14C]palmitate for 3 min; values indicate 14C counts present in organic fractions. Values are means of 2 independent experiments with 3 replicas each and are relative to GFP-infected cells (mean uptake 17.3 fmol palmitate/μg protein/min. C) Lipid synthesis from glucose was measured after incubation of cells with [14C]glucose; values are means ± se of ≥2 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs. GFP-infected cells.
We next assessed the effect of PGC-1 expression on lipid synthesis from glucose. Although insulin tended to increase lipid synthesis, neither PGC-1α or -1β significantly altered lipid synthesis, either in the basal or insulin-stimulated states (Fig. 2C).
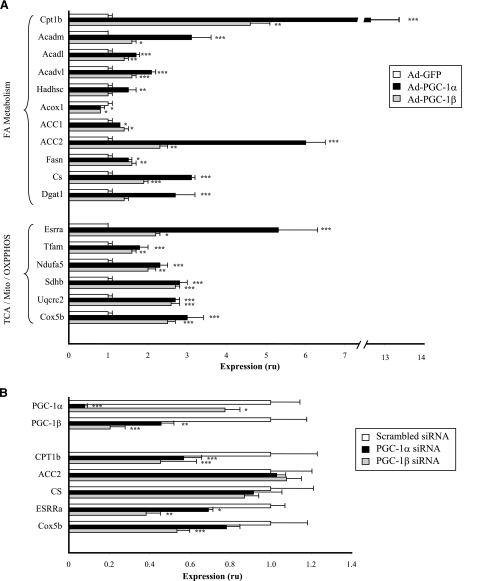
PGC-1 increases mRNA expression of genes critical for both lipid oxidation and synthesis
To identify potential mechanisms mediating the distinct effects of PGC-1α and -1β on lipid metabolism, we examined the mRNA expression of selected genes encoding key enzymes involved in lipid oxidation and synthesis in response to either PGC-1α/β overexpression or siRNA-mediated knockdown (Fig. 3; Supplemental Table 3). Both PGC-1α and -1β markedly increased expression of carnitine palmitoyltransferase 1b (CPT1b) [13.1-fold (P<0.0001) and 4.6-fold (P<0.01) respectively], whereas malonyl-CoA decarboxylase increased by 50% with PGC-1α (P<0.01). Together, these changes would be expected to enhance transport of long-chain fatty acyl-CoAs into the mitochondrial matrix for -oxidation. Similarly, both PGC-1α and -1β significantly increased expression of the medium-, long-, and very long-chain acyl-CoA dehydrogenases (ACADs), critical steps in complete mitochondrial β-oxidation. These effects were greater for PGC-1α than for PGC-1β, ranging from 69 to 213% for PGC-1α (all P<0.01) and from 40 to 63% for PGC-1β (P=0.05 for ACADM and P<0.01 for ACADL and ACADVL). Expression of l-3-hydroxyacyl-CoA dehydrogenase, short chain (HADHSC) was increased in PGC-1α cells by 54 ± 16% (P<0.01) but did not significantly change with PGC-1β. Interestingly, the expression of acyl-CoA oxidase 1, the first enzyme of peroxisomal β-oxidation, was decreased by 22% with PGC-1α (P<0.05) and by 24% with PGC-1β (P< 0.05).
Figure 3.
Effect of PGC-1 overexpression and knockdown on mRNA expression of genes involved in lipid metabolism and mitochondrial biogenesis and function. Total RNA was extracted from C2C12 myotubes infected with GFP, PGC-1α, or PGC-1β adenoviruses (A) or C2C12 myotubes treated with specific PGC-1α or PGC-1β siRNA pools (B). Expression was assessed by quantitative RT-PCR and normalized to TBP levels. Values are means ± se of 1 experiment with 4 replicates and are expressed relative to GFP-infected cells (A) or scrambled siRNA (B). ru, relative units. *P < 0.05, **P < 0.01, ***P < 0.001 vs. respective controls.
We also examined the effects of PGC-1 on mRNA expression of representative genes linked to lipid synthesis (Fig. 3A; Supplemental Table 3). Expression of ACC1 (cytosolic) increased 30 ± 2% (P<0.05) and 39 ± 6% (P<0.01) with PGC-1α and -1β, whereas that of the muscle-dominant mitochondrial ACC2 increased 500 ± 48% (P<0.0001) and 125 ± 15% (P<0.01), respectively. PGC-1α and -1β also increased expression of fatty acid synthase (FAS) (PGC-1α 45±11%, P<0.05; PGC-1β, 57±14%; P<0.01) and citrate synthase (CS) (PGC-1α 212±11%, P<0.0001; PGC-1β 92±5%, P<0.0001). Diacylglycerol O-acyltransferase (DGAT1) increased 170 ± 48% with PGC-1α (P<0.001), with trends to increase with PGC-1β. Thus, PGC-1 expression, particularly that of PGC-1α, increased expression of genes regulating both lipid oxidation and lipogenesis.
PGC-1 overexpression increases expression of genes regulating mitochondrial biogenesis and function
The PGC-1 family of genes regulates gene expression of both nuclear and mitochondrial-encoded genes (39). In agreement, both PGC-1α and -1β expression increased mRNA expression of ESRRα (PGC-1α 425±101% P<0.0001; PGC-1β: 121±10%, P<0.05) and transcription factor A, mitochondrial (TFAM) (PGC-1α 75±17%, P<0.001; PGC-1β 57±6%, P<0.01) (Fig. 3A). Expression of representative genes within complexes I–V of the respiratory chain was significantly increased by both PGC-1α and -1β, generally to a similar extent. This was also reflected by increased mitochondrial protein expression, as assessed by Western blot; for example, PGC-1α increased expression of the NRF-regulated gene aminolevulinic acid synthase by 43 ± 9% (P<0.05) (not shown).
Reduced PGC-1 decreases mRNA expression of genes involved in lipid metabolism
We next assessed the effect of siRNA-mediated knockdown of PGC-1α/β on genes encoding lipid metabolism proteins. As expected, specific siRNA decreased expression of PGC-1α mRNA by 90% (P<0.001) and that of PGC-1β by 50% (P<0.05). In parallel, expression of CPT1b and ESRRα significantly decreased with both PGC-1α and -1β siRNA, and expression of the complex IV Cox5b decreased by 46% with PGC-1β siRNA (P<0.01; similar tendency for PGC-1α siRNA). Interestingly, there were only trends to decreased expression of ACC2 or CS, even with combined PGC-1α/β knockdown, despite the converse increase with PGC-1α/β overexpression.
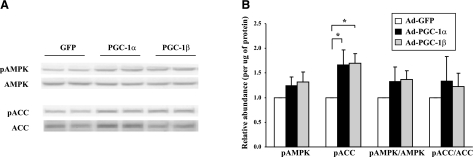
Effects of PGC-1 on cellular AMPK and insulin signaling
AMPK is a key fuel sensor activated by phosphorylation during energy-requiring conditions such as exercise, resulting in (inhibitory) ACC phosphorylation, decreased synthesis of malonyl-CoA, increased fatty acid oxidation, and inhibition of fatty acid synthesis (40). We asked whether the effects of PGC-1 on lipid metabolism might be mediated, in part, by AMPK-dependent pathways. Neither total/phosphorylated AMPK nor total ACC was significantly changed by PGC-1. Interestingly, phosphorylated ACC (inhibited) was increased by almost 70% with both PGC-1α and -1β (P<0.05 for both) (Fig. 4). We observed no major effects of PGC-1α or -1β on insulin signaling.
Figure 4.
Effects of PGC-1 expression on AMPK and ACC protein expression and phosphorylation. Proteins were extracted from C2C12 myotubes at 72 h postinfection, resolved by SDS-PAGE, transferred to 0.45-μm nitrocellulose, and detected using Western blots. A) Representative Western blot for phosphorylated AMPK, total AMPK, phosphorylated ACC, and total ACC. B) Quantification of Western blots. Values are means ± se of 4 independent experiments (2 to 3 replicas each) and are expressed relative to GFP-infected cells. *P < 0.05.
PGC-1 overexpression reduces lipid accumulation, particularly in the serum-starved state
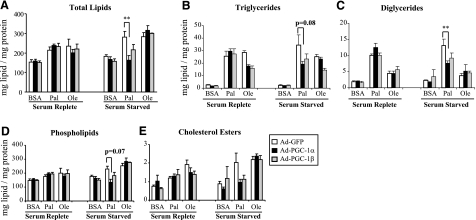
Because PGC-1 overexpression up-regulates expression of genes within both lipid oxidative and synthetic pathways in C2C12 myotubes, we assessed the net effect of PGC-1 expression on lipid content and partitioning at baseline and after exposure to palmitate and oleate, the most prevalent saturated and monounsaturated FFAs in the circulation, respectively (41). As expected, incubation with palmitate or oleate for 24 h tended to increase total lipid content in both serum-replete and serum-starved conditions (vs. BSA-treated cells) (Fig. 5A). Triglycerides were increased by both palmitate and oleate incubation (P<0.05) (Fig. 5B), whereas diglycerides were almost exclusively increased by palmitate (P<0.05) (Fig. 5C). Phospholipids and cholesterol esters tended to increase with palmitate and oleate incubation in most cases (Fig. 5D, E). In the serum-replete state, we observed no significant changes in total lipid content as a function of PGC-1 expression (Fig. 5A, left panel); triglyceride accumulation tended to decrease in oleate-loaded cells with either PGC-1α or -1β (Fig. 5B, left panel).
Figure 5.
Effect of PGC-1 on lipid composition of C2C12 myotubes in serum-replete (left panels) and serum-starved states (right panels). Lipid content and composition was determined in 72-h postinfection C2C12 myotubes incubated in DMEM-H with 2% (serum replete) or 0.1% serum (serum starved) and either BSA, oleate (Ole), or palmitate (Pal) for 24 h. Lipids were extracted with chloroform-methanol (2:1) and separated by thin-layer chromatography. Lipid classes containing fatty acids were also quantitated by GC/MS (as detailed in Materials and Methods). Values are means ± se of ≥2 independent experiments, each with 2 to 4 replicates for all conditions, with the exception of oleate exposure in serum-starved cells (n=1, 4 replicates). Data are expressed as milligrams lipid per milligram protein. A) Total lipids. B) Triglycerides. C) Diglycerides. D) Phospholipids. E) Cholesterol esters. *P < 0.05; **P < 0.01.
In contrast, there were striking differences in PGC-1α effects on lipid content and composition in the serum-starved state. As seen in Fig. 5A, total lipid content in palmitate-loaded cells was decreased by 42 ± 8% (P<0.05) in PGC-1α cells compared with GFP controls. Quantitatively, the majority (82%) of this decrease was accounted for by reductions in the phospholipid fraction (41±9%, P=0.07), which constitutes 69–97% of the lipid content in these cells (depending on conditions). PGC-1α expression also reduced diacylglycerol content by 43% in palmitate-treated cells (P<0.05) (Fig. 5C). The effects of PGC-1β expression were more modest, with a tendency to decreased triglyceride content in cells incubated with oleate (both serum replete and serum starved) or palmitate (serum-starved conditions).
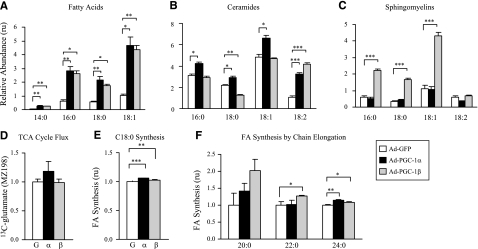
Given the interesting effect of PGC-1 expression on lipid accumulation, we next determined the content of individual fatty acids, ceramides, and sphingomyelins. Fatty acids C14:0, C16:0, C18:0, and C18:1 were increased by 3.1- to 4.6-fold with both PGC-1α and -1β expression in serum-replete C2C12 myotubes (P<0.05 for all vs. GFP control) (Fig. 6A). Similarly, PGC-1α increased C16, C18, C18:1, and C18:2 ceramides; this effect was significant for all chain lengths (P<0.05) but most marked for C18:2 (2.9-fold, P<0.0001). PGC-1β expression was associated with a 42% reduction in C18:0 (P<0.01) and a 3.9-fold increase in C18:2 ceramides (P<0.001), compared with those for GFP (Fig. 6B). PGC-1α did not significantly alter C16, C18, C18:1, or C18:2 sphingomyelins. However, PGC-1β increased sphingomyelin C16, C18, and C18:1 by 3.7- to 5.1-fold (P<0.001 for all), with no significant effect on C18:2 (Fig. 6C).
Figure 6.
Effect of PGC-1 on intracellular lipid flux and synthesis after incubation with [13C]glucose or [13C]stearate (18:0). C2C12 myotubes were incubated at 24 h postinfection with 5 mM glucose-5 mM [d-1,2-13C2]glucose (A–C) or 1% BSA-125 μM [13C]stearic acid (D–F) in DMEM-H, with 2% HS for 72 h. A–C) Relative contents of fatty acids (A), ceramides (B), and sphingomyelins (C) were determined by SIDMAP and GC/MS. D) 13C incorporation into glutamate (MZ198) was measured as an indicator of TCA cycle flux. E) Synthesis of C18 by chain shortening and elongation from [13C]stearate. F) Synthesis of C20, C22, and C24 by chain elongation from [13C]stearate (C18). Values are means ± se. Values are expressed relative to GFP-infected cells in panels D–F. *P < 0.05; **P < 0.01; ***P < 0.001. Note that some error bars are too small to be visualized.
Metabolomic analysis of C2C12 myotubes overexpressing PGC-1
To determine the specific steps in carbohydrate and lipid metabolism influenced by PGC-1 expression and potentially responsible for alterations in lipid content, we performed SIDMAP. C2C12 myotubes were incubated with [13C]glucose or [13C]stearate for 72 h, beginning at 24 h after adenoviral infection, and incorporation of 13C into metabolites was determined. No 13C tracer was found in secreted lactate or ribose when cells were treated with [13C]stearate (not shown). However, when glucose was used as tracer, we observed 24.2 to 26.3% 13C-labeled lactate fractions (Σm) in the medium of all groups, of which >95% was produced via direct glycolysis with no apparent difference in response to PGC-1 expression status. There was no significant change in labeled glutamate (m/z 198), indicating no major differences in the tricarboxylic acid (TCA) cycle anaplerotic flux (Fig. 6D). There was a modest, but consistent, trend for increased de novo synthesis from glucose; these reached statistical significance for stearic acid (C18:0) 13C enrichment (Σm and acetyl-CoA 13C enrichment, as well as for fractional new synthesis in PGC-1α-expressing cells; P<0.05 for all).
We next examined the synthesis of individual fatty acids in cells incubated with [13C]stearate. C18 synthesis, occurring by chain shortening to C16 and then elongation back to C18 (futile cycle) was increased by 6 and 2% with PGC-1α (P<0.001) and PGC-1β (P<0.01), respectively (Fig. 6E). More strikingly, synthesis of long-chain fatty acids, occurring by chain elongation, increased with PGC-1α and -1β, reaching significance for C22 synthesis with PGC-1β (27±1% increase; P=0.05) and for C24 synthesis with both PGC-1α and -1β (14±2 and 9±1% increase, respectively; P<0.05 for both) (Fig. 6F).
DISCUSSION
Investigations in transgenic and null animals have emphasized the critical role for PGC-1 family genes as modulators of systemic metabolism (7, 10, 14, 16, 17). However, it remains unclear whether reductions in PGC-1α and -1β expression and/or transcriptional activity may contribute to alterations in oxidative capacity and increased lipid accumulation associated with human insulin resistance. Thus, to define the complex effects of PGC-1 on lipid metabolism, we overexpressed PGC-1α and -1β or GFP adenovirus in C2C12 myotubes. We recognize that the sustained high-level expression (8.5-fold greater than skeletal muscle for PGC-1α protein) may produce effects that differ from those observed in vivo, when levels are typically modulated by physiological stimuli (e.g., exercise). Nevertheless, C2C12 myotubes express relatively low levels of PGC-1 compared with in vivo levels (13) and, thus, are a good model in which to study the effect of PGC-1 on cellular metabolic flux. We now demonstrate that PGC-1α and -1β overexpression causes a modest but significant increase in both glucose and fatty acid oxidation, as expected, and also reveal an unexpected PGC-1-linked increase in lipid synthesis and fatty acid content and altered lipid composition.
The modest increase in glucose oxidation induced by PGC-1 expression may be mediated at several levels, including glucose uptake, glycolysis, or downstream oxidative pathways. We did observe a stimulatory effect of PGC-1α on both basal and insulin-stimulated deoxyglucose uptake and mRNA expression of the insulin-responsive GLUT4 glucose transporter after short-term serum starvation, in agreement with the results of Michael et al. (13) and the increase in basal glucose transport in mice with muscle-specific PGC-1α expression (12). Increased glucose oxidation may also reflect flux through glycolysis, as suggested by increased secretion of lactate into the medium with PGC-1α (Fig. 1D). SIDMAP analysis in palmitate-labeled cells indicated no significant alteration in glutamate pool labeling and thus unchanged TCA cycle flux. Thus, we hypothesize that increased glycolytic flux is responsible for increased lactate secretion, but that complete glucose oxidation is limited by reduced entry of pyruvate into the TCA cycle, perhaps mediated by pyruvate dehydrogenase kinase 4 repression of pyruvate dehydrogenase (12).
Disordered lipid oxidation is a common feature of both insulin resistance and type 2 diabetes (42) and may contribute to intramyocellular lipid accumulation and inhibition of insulin action (43). Consistent with our original hypothesis, we observed a significant 25–30% increase in fatty acid oxidation as a function of PGC-1 (Fig. 2A). Our data are also consistent with those of Koves et al. (29), who demonstrated that PGC-1α expression can increase complete β-oxidation of lipids, thus contributing to beneficial effects of exercise training on oxidative function.
To identify potential transcriptional changes contributing to enhanced lipid oxidation, we examined mRNA expression of genes regulating lipid metabolism as a function of PGC-1 expression. Interestingly, PCR analysis suggests dual effects of PGC-1 to modulate expression of genes regulating both lipid catabolism and synthesis. As predicted, PGC-1 overexpression increased mRNA expression of genes encoding enzymes of critical steps of lipid oxidation (e.g., CPT1b, HADHSC, ACADM, ACADL, and ACADVL); regulatory genes critical for mitochondrial biogenesis and function, including ERRα and TFAM; and nuclear genes encoding mitochondrial oxidative phosphorylation complex proteins (Fig. 3; Supplemental Table 3). Together, these changes would be predicted to enhance mitochondrial oxidative capacity. In parallel, however, PGC-1 also increased mRNA expression of genes promoting lipid synthesis (e.g., ACC, FAS, CS, and DGAT1), potentially contributing to the observed net increase in fatty acid content (Fig. 6A). Thus, PGC-1 expression results in both increased fatty acid oxidation and lipid synthesis, suggesting a net increase in turnover of lipids.
Interestingly, siRNA-mediated knockdown of either PGC-1α or -1β reduced expression of representative lipid oxidation and OXPHOS genes (e.g., CPT1, ERRα, and COX5b) but had no significant effect on ACC2 or CS (even with combined knockdown of PGC-1α/β). We hypothesize that these differential effects on specific gene targets could potentially result from differential effects on autoregulatory transcriptional loops involving PGC-1, GABPA, and ERR proteins (4). In this setting, siRNA-mediated reduction in PGC-1α and/or -1β expression reduces mRNA expression of ERRα, interrupting the PGC-1/ERR double-positive feedback loop and therefore decreasing expression of those genes, which are both PGC-1- and ERRα-dependent (e.g., ERRα, COX5b, and CPT1b) but not of ERR-independent genes (4, 44, 45). Alterations in PGC-1 acetylation or localization differentially affected by adenoviral overexpression vs. siRNA targeting could also affect expression of pools of PGC-1-responsive genes.
Because metabolic flux is determined not only by gene or protein expression alone but also by substrate concentration, post-translational modification of enzymes, and allosteric modulation, we determined the net effect of PGC-1 expression on total lipid, lipid fractions, and flux. PGC-1α/β overexpression altered lipid content and composition, particularly in serum-starved conditions. Total lipid content in palmitate-treated cells was decreased by 42% in cells expressing PGC-1α, a finding largely accounted for by the reduction in phospholipid content. Diacylglycerol content was also significantly reduced by PGC-1α expression in palmitate-loaded cells (Fig. 5C). This is of particular interest given the deleterious effects of lipid-induced diacylglycerols and protein kinase C activation on insulin sensitivity (46,47,48). Although the effects on both total lipid and triglycerides are dominant in the serum-starved state, triglyceride content tended to decrease in oleate-loaded cells even in the serum-replete state (Fig. 5B).
To determine the specific steps in carbohydrate and lipid metabolism affected by PGC-1 expression, we performed SIDMAP studies, demonstrating a very significant effect of PGC-1α or -1β to increase C14:0, C16:0, C18:0, and C18:1 content after glucose exposure (Fig. 6). These data also parallel the trend to increased fatty acid synthesis from [14C]glucose during short-term (1 h) incubations (Fig. 2C); the longer-term SIDMAP studies suggest that differences in fatty acid synthesis may indeed have significant effect on lipid accumulation over time. Moreover, fatty acid synthesis for long-chain fatty acids (by chain elongation) increased with overexpression of either PGC-1α or -1β, reaching significance for C22 and C24. The mechanisms for elongation are not clear at this time; mRNA expression of Elovl1 and Elovl5, the mammalian elongases detected in C2C12 myotubes, did not significantly change with PGC-1α and tended to decrease with PGC-1β (not shown). Although activity of elongases is typically regulated at a transcriptional level, we cannot exclude changes in activity, substrate flux, or subcellular localization (49).
Ceramides have emerged as key mediators of insulin resistance induced by saturated lipids (38), and plasma ceramides are associated with insulin resistance in humans (40, 50). Conversely, insulin action, body weight, and energy expenditure can be improved with inhibition of ceramide synthesis (51). Interestingly, ceramide content (C16, C18, C18:1, and C18:2) was increased by PGC-1α-expressing cells, whereas PGC-1β did not change C16 or C18:1, decreased 18:0, and increased 18:2. On the other hand, PGC-1α did not change sphingomyelin content, whereas PGC-1β increased C16, C18, and C18:1. Although the mechanisms responsible for these patterns remain unclear, it has recently been demonstrated that specific ceramide species (with distinct fatty acid moieties) can be regulated independently by the multiple members of the ceramide synthase/longevity assurance genes (LAG family) (41) and that different species may contribute to distinct effects on cellular stress responses (52). Thus, we speculate that alterations in ceramide subspecies or reductions in ceramide/sphingomyelin ratios induced by PGC-1β may also be beneficial for metabolic homeostasis; further studies will be required to evaluate mechanisms mediating differential effects on ceramide species.
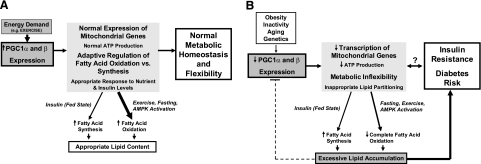
Taken together, our data suggest that PGC-1α and -1β modulate lipid metabolism in myotubes, facilitating both lipid synthesis and storage, thus serving as primary modulators of lipid fuel availability and utilization. In the serum-replete state, the net effect of PGC-1-mediated transcriptional changes is to enhance energy storage via increased lipid synthesis from glucose; in the serum-starved state, PGC-1 promotes lipid oxidation (Fig. 7A).
Figure 7.
Proposed model of role of PGC-1 in lipid metabolism. Our results suggest that PGC-1 is a critical modulator of lipid metabolism, content, and composition, which regulates appropriate lipid content to ensure adequate ATP production and sufficient fuel availability for energy-requiring states (e.g., exercise). A) With exercise, PGC-1 mRNA and protein expression are increased, resulting in increased expression of genes involved in both mitochondrial biogenesis and lipid metabolism. Expression of genes favoring fatty acid oxidation is dominant in this setting; in contrast, in the fed state, fatty acid synthesis may be dominant, promoting storage of lipid for subsequent bouts of exercise. B) Reductions in PGC-1 expression or function, caused by obesity, chronic inactivity, aging, or genetic predisposition, may contribute to a vicious cycle, promoting impairment of both mitochondrial oxidative function and lipid utilization, leading to reductions in capacity for complete oxidation of lipids and inappropriate lipid accumulation. In turn, sustained lipid accumulation may further reduce PGC-1 expression, perpetuating a vicious cycle of reduced oxidative capacity associated with both insulin resistance and diabetes.
Our data are in agreement with results from transgenic mice expressing PGC-1α, in which increases in both mRNA and protein expression of OXPHOS and fatty acid oxidation genes are observed, in parallel with increased ATP synthesis, fatty acid oxidation, and exercise performance (53). Interestingly, when these mice were fed a high-fat diet, increased lipid accumulation in skeletal muscle was observed, with decreased insulin sensitivity. Thus, high-level PGC-1 expression can increase expression of genes promoting both fatty acid oxidation and fatty acid/triglyceride synthesis, as observed in the present study. If this expression is not paralleled by lipid oxidation (as with exercise-induced fuel utilization) or the ability to modulate PGC-1 expression appropriately, accumulation of lipids and/or incomplete oxidation products may indeed contribute to reduced insulin sensitivity.
Because exercise is a key regulator of PGC-1 expression in vivo (54), we speculate that the net physiological effects of PGC-1-mediated transcriptional changes are to prepare muscle for subsequent bouts of exercise by both facilitating lipid storage and enhancing oxidative capacity. In the setting of acute exercise, when insulin levels are low and PGC-1 is robustly induced, or in the fasting state, lipid stores can be effectively oxidized for energy needs. This perspective also suggests that increased lipid accumulation in the trained athlete (55) is actually a physiologically appropriate condition that may be mediated by repetitive exercise-induced increases in protein expression of PGC-1. Indeed, the capacity of muscle mitochondria to fully oxidize a heavy influx of fatty acids is correlated with PGC-1α mRNA expression levels in both rodent muscle and L6 myotubes (29). Modest transgenic overexpression of PGC-1α in mice also is associated with increased insulin sensitivity and palmitate oxidation in subsarcolemmal mitochondria, in parallel with up-regulation of substrate transport proteins (GLUT4, FABPpm, and FAT/CD36) and oxidative mitochondrial proteins (14). Our data, including gene expression data, lipid composition, and SIDMAP analysis further extend the concept that PGC-1α expression may enhance the lipid-induced substrate switch in myotubes (29), facilitating both lipid storage and oxidation, depending on the ambient metabolic/hormonal milieu (Fig. 7A).
Our data can be interpreted more broadly, given that alterations in PGC-1 expression and/or activity may also contribute to pathophysiology of skeletal muscle insulin resistance and “metabolic inflexibility” associated with obesity and diabetes. Both chronic insulin resistance and diabetes are accompanied by impaired oxidative capacity and “ectopic” lipid accumulation in muscle (33, 56, 57), including pathogenic ceramides and diacylglycerol lipid intermediates, among others (45). In both humans and in cultured myotubes, experimental lipid loading is associated with down-regulation of PGC-1 mRNA expression (28, 30, 1). Such changes may be viewed as an initially appropriate negative feedback loop, serving to limit further lipogenesis and lipid accumulation in the setting of lipid nutrient excess. With prolonged inactivity or continued nutrient excess, fuel utilization via lipid oxidation is limited, lipid accumulation persists, and decreased PGC-1 mRNA expression is sustained (Fig. 7B). This may contribute to a vicious cycle, further impairing oxidative mitochondrial function and leading to increased production of lipid intermediates associated with insulin resistance.
Supplementary Material
Acknowledgments
The authors thank Dr. Bruce Spiegelman (Dana Farber Cancer Institute, Boston, MA, USA) for the gift of adenovirus constructs Ad-GFP, AdPGC-1α, and AdPGC-1β and Drs. Masa Katic, Yuji Yamamoto, and Enxuan Jing (Joslin Diabetes Center, Boston, MA, USA) for selected primers. The authors appreciate the assistance of the Lipid Core, Mouse Metabolic Phenotyping Center, Vanderbilt University (Nashville, TN, USA). The authors gratefully acknowledge grant funding from the National Institute of Diabetes and Digestive and Kidney Diseases (DK062948) and the Diabetes and Endocrinology Research Center (P30 DK36836).
References
- Vega R B, Huss J M, Kelly D P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon J C, Puigserver P, Fan M, Gonzalez F J, Spiegelman B M. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R C. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- Mootha V K, Handschin C, Arlow D, Xie X, St. Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy P J, Schulman I G, Heyman R A, Lander E S, Spiegelman B M. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yang R, Tarr P T, Wu P H, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, Newgard C B, Spiegelman B M. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr P T, Zhang C Y, Wu Z, Boss O, Michael L F, Puigserver P, Isotani E, Olson E N, Lowell B B, Bassel-Duby R, Spiegelman B M. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman B M. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey C J, Yoon J C, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman B M. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr P T, Spiegelman B M. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N, Kawada T, Miyoshi M, Ezaki O, Kakizuka A. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Wende A R, Schaeffer P J, Parker G J, Zechner C, Han D H, Chen M M, Hancock C R, Lehman J J, Huss J M, McClain D A, Holloszy J O, Kelly D P. A role for the transcriptional coactivator PGC-1α in muscle refueling. J Biol Chem. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- Michael L F, Wu Z, Cheatham R B, Puigserver P, Adelmant G, Lehman J J, Kelly D P, Spiegelman B M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton C R, Nickerson J G, Lally J, Han X X, Holloway G P, Glatz J F, Luiken J J, Graham T E, Heikkila J J, Bonen A. Modest PGC-1α overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem. 2008;283:4228–4240. doi: 10.1074/jbc.M704332200. [DOI] [PubMed] [Google Scholar]
- Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu P H, Rybkin I, Shelton J M, Manieri M, Cinti S, Schoen F J, Bassel-Duby R, Rosenzweig A, Ingwall J S, Spiegelman B M. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu P H, Tarr P T, Lindenberg K S, St. Pierre J, Zhang C Y, Mootha V K, Jager S, Vianna C R, Reznick R M, Cui L, Manieri M, Donovan M X, Wu Z, Cooper M P, Fan M C, Rohas L M, Zavacki A M, Cinti S, Shulman G I, Lowell B B, Krainc D, Spiegelman B M. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Leone T C, Lehman J J, Finck B N, Schaeffer P J, Wende A R, Boudina S, Courtois M, Wozniak D F, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy J O, Medeiros D M, Schmidt R E, Saffitz J E, Abel E D, Semenkovich C F, Kelly D P. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur N K, Yan Z, Spiegelman B M. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1α muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- Handschin C, Choi C S, Chin S, Kim S, Kawamori D, Kurpad A J, Neubauer N, Hu J, Mootha V K, Kim Y B, Kulkarni R N, Shulman G I, Spiegelman B M. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1α knockout mice reveals skeletal muscle-pancreatic β cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelliott C J, Medina-Gomez G, Petrovic N, Kis A, Feldmann H M, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin S E, Zaha V G, Fountain K T, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly Y, Storlien L, Stromstedt M, Snaith M, Oresic M, Abel E D, Cannon B, Vidal-Puig A. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianna C R, Huntgeburth M, Coppari R, Choi C S, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim Y B, Cinti S, Shulman G I, Spiegelman B M, Lowell B B. Hypomorphic mutation of PGC-1β causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 2006;4:453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Mehl I, Chong L-W, Nofsinger R, Evans R. PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci U S A. 104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti M E, Butte A J, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker E J, Goldfine A B, Mun E, DeFronzo R, Finlayson J, Kahn C R, Mandarino L J. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V K, Lindgren C M, Eriksson K F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly M J, Patterson N, Mesirov J P, Golub T R, Tamayo P, Spiegelman B, Lander E S, Hirschhorn J N, Altshuler D, Groop L C. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Larrouy D, Vidal H, Andreelli F, Laville M, Langin D. Cloning and mRNA tissue distribution of human PPARγ coactivator-1. Int J Obes Relat Metab Disord. 1999;23:1327–1332. doi: 10.1038/sj.ijo.0801106. [DOI] [PubMed] [Google Scholar]
- Hammarstedt A, Jansson P A, Wesslau C, Yang X, Smith U. Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochem Biophys Res Commun. 2003;301:578–582. doi: 10.1016/s0006-291x(03)00014-7. [DOI] [PubMed] [Google Scholar]
- Semple R K, Crowley V C, Sewter C P, Laudes M, Christodoulides C, Considine R V, Vidal-Puig A, O'Rahilly S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1α is reduced in the adipose tissue of morbidly obese subjects. Int J Obes Relat Metab Disord. 2004;28:176–179. doi: 10.1038/sj.ijo.0802482. [DOI] [PubMed] [Google Scholar]
- Crunkhorn S, Dearie F, Mantzoros C, Gami H, da Silva W S, Espinoza D, Faucette R, Barry K, Bianco A C, Patti M E. Peroxisome proliferator activator receptor γ coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem. 2007;282:15439–15450. doi: 10.1074/jbc.M611214200. [DOI] [PubMed] [Google Scholar]
- Koves T R, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm G L, Yan Z, Newgard C B, Muoio D M. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280:33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- Sparks L M, Xie H, Koza R A, Mynatt R, Hulver M W, Bray G A, Smith S R. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- Richardson D K, Kashyap S, Bajaj M, Cusi K, Mandarino S J, Finlayson J, DeFronzo R A, Jenkinson C P, Mandarino L J. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases expression of extracellular matrix genes in human skeletal muscle. J Biol Chem. 2004;280:10290–10297. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- Kelley D E, He J, Menshikova E V, Ritov V B. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- Petersen K F, Dufour S, Befroy D, Garcia R, Shulman G I. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella L L, Pilch P F. C2C12 myocytes lack an insulin-responsive vesicular compartment despite dexamethasone-induced GLUT4 expression. Am J Physiol Endocrinol Metab. 2002;283:E514–E524. doi: 10.1152/ajpendo.00092.2002. [DOI] [PubMed] [Google Scholar]
- Folli F, Saad M J A, Backer J M, Kahn C R. Regulation of phosphatidylinositol 3-kinase activity in liver and muscle of animal models of insulin-resistant and insulin-deficient diabetes mellitus. J Clin Invest. 1993;92:1787–1794. doi: 10.1172/JCI116768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babson A L, Shapiro P O, Phillips G E. A new assay for cholesterol and cholesterol esters in serum which is not affected by bilirubin. Clin Chim Acta. 1962;7:800–804. doi: 10.1016/0009-8981(62)90062-1. [DOI] [PubMed] [Google Scholar]
- Bodennec J, Koul O, Aguado I, Brichon G, Zwingelstein G, Portoukalian J. A procedure for fractionation of sphingolipid classes by solid-phase extraction on aminopropyl cartridges. J Lipid Res. 2000;41:1524–1531. [PubMed] [Google Scholar]
- Storey J D. A direct approach to false discovery rates. J R Stat Soc B. 2002;64:479–498. [Google Scholar]
- Holland W L, Brozinick J T, Wang L P, Hawkins E D, Sargent K M, Liu Y, Narra K, Hoehn K L, Knotts T A, Siesky A, Nelson D H, Karathanasis S K, Fontenot G K, Birnbaum M J, Summers S A. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5:167–179. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Adams J M, Pratipanawatr T, Berria R, Wang E, DeFronzo R A, Sullards M C, Mandarino L J. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes. 2004;53:25–31. doi: 10.2337/diabetes.53.1.25. [DOI] [PubMed] [Google Scholar]
- Pewzner-Jung Y, Ben Dor S, Futerman A H. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- Simoneau J A, Kelley D E. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol. 1997;83:166–171. doi: 10.1152/jappl.1997.83.1.166. [DOI] [PubMed] [Google Scholar]
- Kelley D E, Mandarino L J. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49:677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- Dufour C R, Wilson B J, Huss J M, Kelly D P, Alaynick W A, Downes M, Evans R M, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Schreiber S N, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- Montell E, Turini M, Marotta M, Roberts M, Noe V, Ciudad C J, Mace K, Gomez-Foix A M. DAG accumulation from saturated fatty acids desensitizes insulin stimulation of glucose uptake in muscle cells. Am J Physiol Endocrinol Metab. 2001;280:E229–E237. doi: 10.1152/ajpendo.2001.280.2.E229. [DOI] [PubMed] [Google Scholar]
- Itani S I, Ruderman N B, Schmieder F, Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IκB-α. Diabetes. 2002;51:2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- Savage D B, Petersen K F, Shulman G I. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev. 2007;87:507–520. doi: 10.1152/physrev.00024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Haus J M, Kashyap S R, Kasumov T, Zhang R, Kelly K R, DeFronzo R A, Kirwan J P. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 2009;58:337–343. doi: 10.2337/db08-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Badeanlou L, Bielawski J, Roberts A J, Hannun Y A, Samad F. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. Am J Physiol Endocrinol Metab. 2009;297:E211–E224. doi: 10.1152/ajpendo.91014.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder C M. Cell biology. Ceramides—friend or foe in hypoxia? Science. 2009;324:343–344. doi: 10.1126/science.1173278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo J A, Daniels T G, Wang X, Paul A, Lin J, Spiegelman B M, Stevenson S C, Rangwala S M. Muscle-specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- Russell A P, Hesselink M K, Lo S K, Schrauwen P. Regulation of metabolic transcriptional co-activators and transcription factors with acute exercise. FASEB J. 2005;19:986–988. doi: 10.1096/fj.04-3168fje. [DOI] [PubMed] [Google Scholar]
- Goodpaster B H, He J, Watkins S, Kelley D E. Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. J Clin Endocrinol Metab. 2001;86:5755–5561. doi: 10.1210/jcem.86.12.8075. [DOI] [PubMed] [Google Scholar]
- Jacob S, Machann J, Rett K, Brechtel K, Volk A, Renn W, Maerker E, Matthaei S, Schick F, Claussen C D, Haring H U. Association of increased intramyocellular lipid content with insulin resistance in lean nondiabetic offspring of type 2 diabetic subjects. Diabetes. 1999;48:1113–1119. doi: 10.2337/diabetes.48.5.1113. [DOI] [PubMed] [Google Scholar]
- Simoneau J A, Kelley D E. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol. 1997;83:166–171. doi: 10.1152/jappl.1997.83.1.166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.