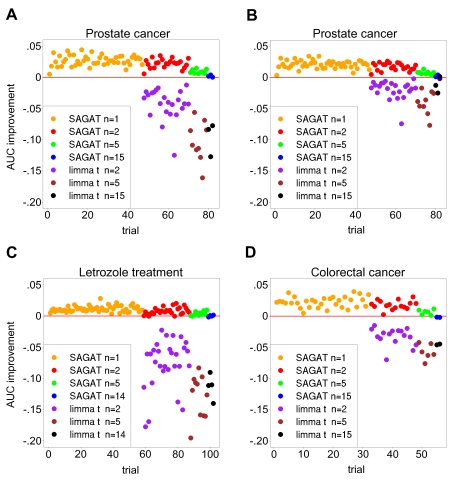
Figure 5. SAGAT, fold change, and limma t performance on subsets of three highly replicated human datasets.
In each panel, the gold standard was defined as the top 1122, 588, and 6002 highest scoring genes for the prostate cancer, letrozole treatment, and colorectal cancer datasets, respectively. (A) The fold change metric was used to rank genes for the gold standard. SAGAT (with HGU95Av2 compendium), fold change, and limma t were run on all combinations of n = 1, 2, 5, and 15 replicate subsets and the performance improvement of SAGAT and limma t over fold change displayed. Limma t requires two or more replicates to score genes. (B) Identical conditions as (A), with the limma t metric used to rank gold standard genes. (C) and (D) Similar plots for the letrozole treatment (HGU133A compendium) and colorectal cancer (HGU133plus2.0 compendium) datasets, respectively. Fold change was used to rank gold standard genes in these panels. In all cases, SAGAT performs as well or better than the fold change and limma t metrics, while the limma t nearly always yields the poorest performance.