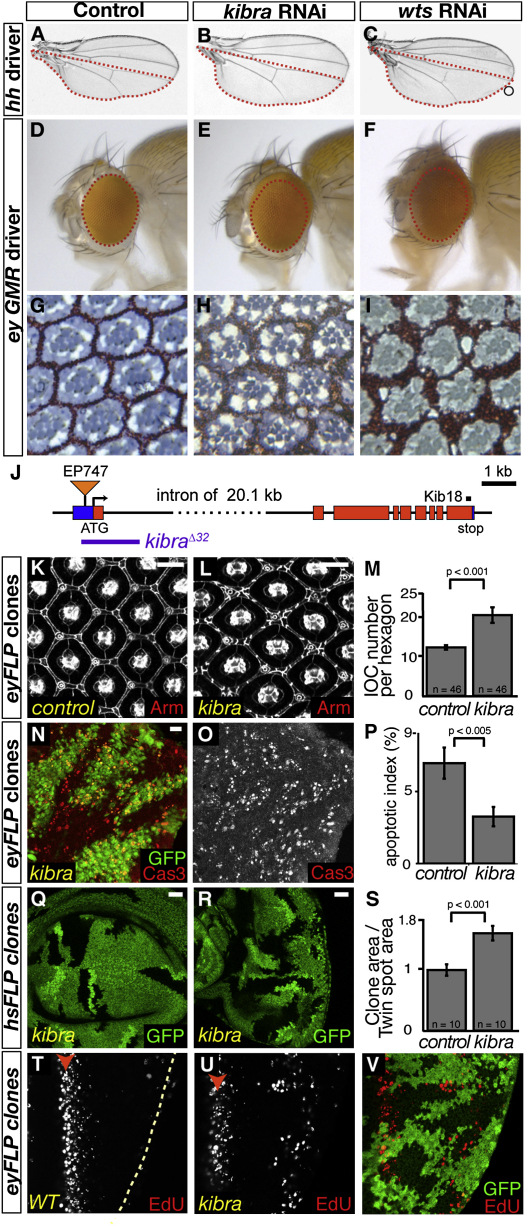
Figure 1.
Kibra Loss of Function Induces a Phenotype Similar to SWH Network Loss of Function
(A–C) Control wings (A) and wings where the kibra (B) or wts (C) genes were silenced by RNAi in the posterior compartment (red dotted line).
(D–I) Control fly eyes (D and G) and eyes expressing the same RNAi lines against kibra (E and H) or wts (F and I). In adult eye sections (G–I), interommatidial cells (IOCs) are in red and photoreceptors in blue.
(J) Schematic of the kibra locus showing the localization of the kibraΔ32 allele, generated by excision of the EP747 P element (triangle). The coding sequence is in red; 5′- and 3′-UTRs are in blue. In black is the peptide recognized by the Kib18 antibody.
(K–M) 40 hr APF retinas containing kibra mutant clones. (K and L) Cell outlines are visualized by anti-Arm staining (scale bar = 10 μm). (M) Quantification of the IOCs. The p value from a Mann-Whitney test is shown.
(N–P) Apoptotic index in 28 hr APF retinas containing kibra mutant clones (absence of GFP). A retina stained for activated Caspase-3 (Cas3) merged with GFP is shown in (N). The Cas3 staining for the whole retina is shown in (O). (P) Quantification of the apoptotic index. The p value from a Mann-Whitney test is shown.
(Q–S) Wing disc (Q) and eye disc (R) containing kibra clones (lack of GFP). Scale bars = 20 μm. (S) Quantification of the proliferation advantage of WT or kibra mutant cells by calculating the ratio between total clonal area (no GFP) and total twin spot area (two copies of GFP) for discs containing either WT or kibra clones. The p value from a Mann-Whitney test is shown.
(T–V) EdU labeling (shown in gray or red) of WT discs (T) and of discs containing kibra clones (U and V). Posterior is to the right, and the SMW is indicated by a red arrowhead. See also Figure S1.
Error bars represent standard deviations.