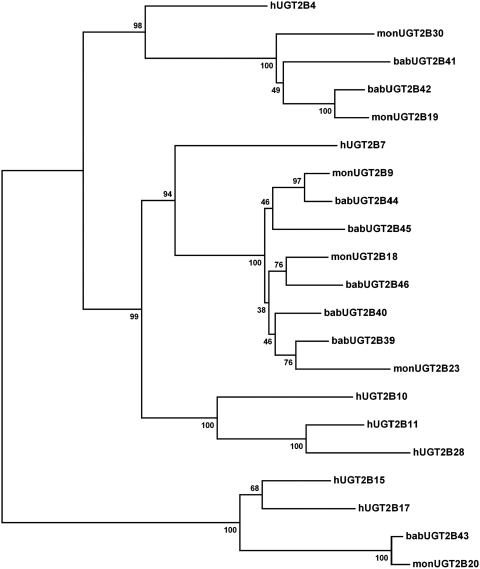
Fig. 1.
Neighbor-joining (NJ) reconciled phylogenetic tree with baboon, human, and monkey (M. fascicularis) UGT2B enzymes. The sequences for human and monkey UGT2B genes were obtained from the GenBank. The protein coding regions were aligned using MEGA4.0, and an NJ distance-based phylogenetic tree using p distances was generated. Robustness of bifurcations is estimated by bootstrap analysis with 1000 replicates, and the consensus tree is shown. Numbers at bifurcations indicate bootstrap values in percent. Annotation: bab (baboon, P. anubis), h (human), and mon (cynomolgus monkey, M. fascicularis).