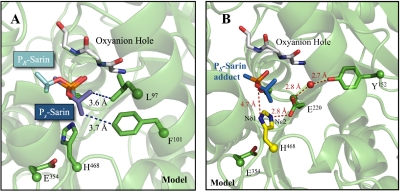
Fig. 7.
hCE1-sarin model. A, PR and PS enantiomers of sarin modeled in the hCE1 active site. The O-isopropyl group on PS sarin makes hydrophobic contacts with Phe101 and Leu97, whereas PR does not have any additional interactions. B, model of proposed mechanism of PS sarin reactivation in hCE1. In AChE, His468 has been observed to rotate away from Glu354 and interact with Glu221 after acylation. Modeling this shift in the hCE1 active site, there is an electronic network formed between Tyr152 (green) and His468 (yellow) that may either deprotonate Nε2 or allow Nδ1 of His468 to act as a general base for water activation. PS sarin (blue) was modeled into the hCE1-soman structure (RCSB PDB access code 2hrq; Fleming et al., 2007).