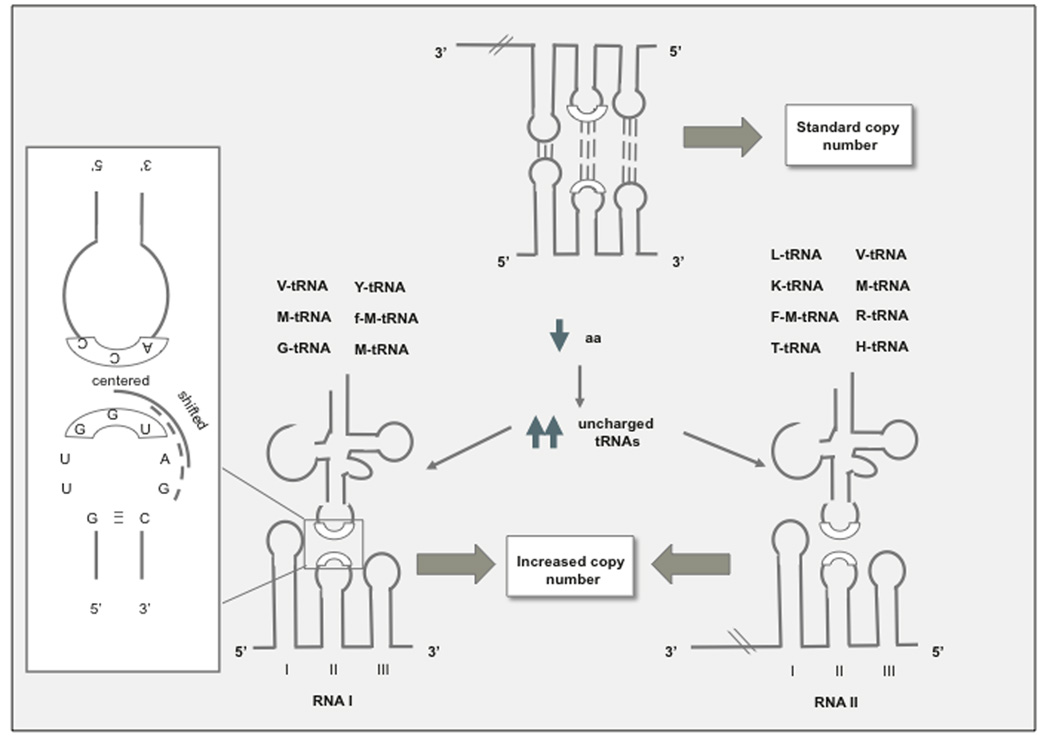
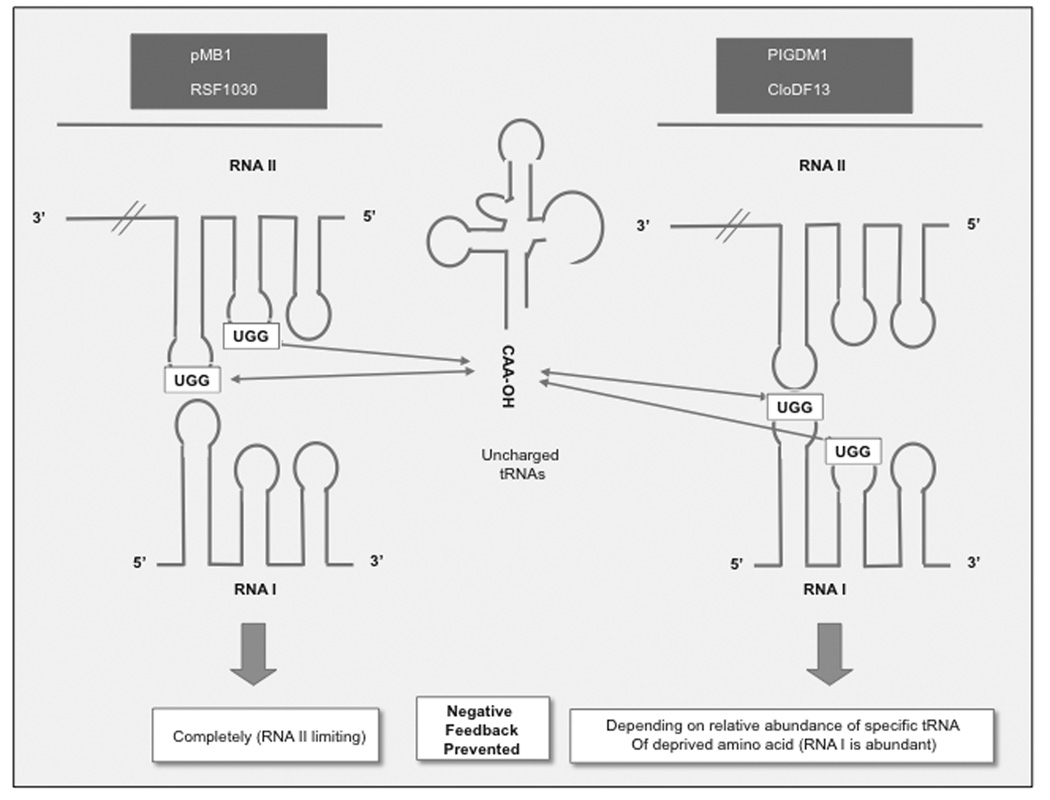
Fig. (4). Models of disregulation of plasmid replication caused by accumulation of uncharged tRNAs in relA stains.
a Formation of codon-anticodon complexes with tRNA. A structural similarity and >40% sequence homology was noticed between SL1, 2, and 3 of RNA I, II or both and the cloverleaf structure of t-RNAs. Yavachev et al. postulated that competitive hybridization between the anticodon loop of tRNA and the corresponding anticodon-like loops of RNA I or RNA II could interfere with the formation of RNAI/RNAII hybrids [36]. This model predicts that changes in plasmid copy number with deprivation of individual amino acids should correlate with the homology between the corresponding tRNAs and loops in RNA I and II. Note that each unpaired loop provides three different options for hybridization with tRNA anticodon sequences. A 7-nt loop is shown in the inset as an example: centered (boxed), shifted (continuous line) and very shifted (broken line). Based on solvent exposure, centered interactions are assumed to be the preferred ones, while the very shifted ones would be the least preferred ones. b. CAA-OH tRNA hybridization with UGG sequence of RNAI or RNA II. 3’-CAA of uncharged tRNAs hybridizes with the GGU motif of either RNA I or RNAII (depending on the specific sequence of each ori), and this bond is stabilized because a proton given by the CCA is trapped by an electron hole (GG+) at RNA I, RNA II loops [37]. The specific effects of individual amino acid deprivation are explained because in ColE1 plasmids, the GGU…G sequence is encoded in RNA I, which is the more abundant of the two ori transcripts and therefore sensitive to the levels of uncharged tRNA, which in turn corresponds to the relative abundance of different amino acids in E. coli proteins. This model predicts that starvation for any amino acid should lead to runaway plasmid replication in the case of PIGDM1, CloDF13 and other members of the ColE1/pMB1 family of enzymes that encode the GGU…G in the RNAII transcript. Note that both models propose a role of tRNAs in regulation of replication initiation. It has been proposed that tRNAs originated from RNA replication primers not unlike RNA II, as clover leaf-like structures frequently prime replication of viral and plasmid genomes (note the clover leaf-like structure of RNA II) [88].