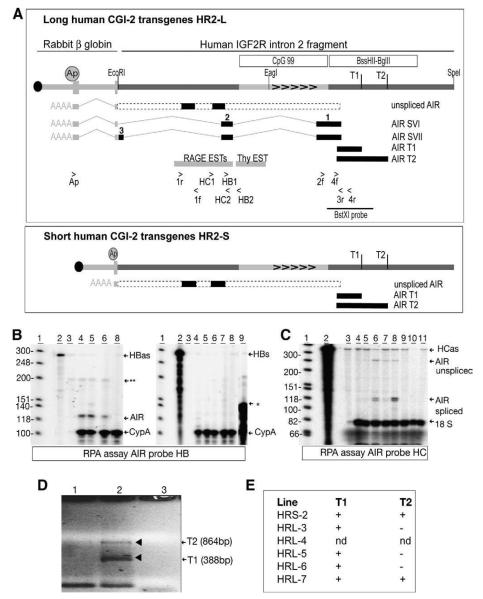
Fig. 3.
Human CGI-2 transgenes express spliced and unspliced transcripts. (A) Human CGI-2 transcripts analyzed by RT-PCR for HR2-L (top box) and HR2-S (lower box) transgenes. A transgene map is shown above (details as in Fig. 2A) with primer locations. T1, T2 indicate the sequence identified by 5′RACE with primers 2f+4f, as transcriptional start sites (D). Grey bars indicate ESTs in this region (http://genome.ucsc.edu) see Sup. Fig. 2. The position of splice variants SVI and SVII identified by primers Ap+3r, is shown. Introns are indicated by grey lines, exons by numbered black bars. The dashed bar indicates the predicted unspliced AIR transcript corresponding to the 4 kb Northern blot signal in Fig. 2C. Black bars within this indicate unspliced AIR transcripts detected with primers 1r, 1f and HC RPA probe. HC1/HC2 and HB1/HB2: primers used to prepare RNase protection probes. (B) RNase protection assay with probe HB in brain samples from adult transgenic mice showing expression of spliced human CGI-2 transcripts. The probe HBas protects transcripts with the opposite orientation to IGF2R and identifies a 120 bp spliced SVI and SVII transcript (labeled AIR, present in lanes 4–6), and a 199 bp unspliced transcript (not distinguishable from a non-specific product**). Probe HBs protect a 120 bp product with the same orientation to IGF2R and fails to identify any transcripts from the human CGI-2. An in vitro transcribed 140 bp fragment from probe HBas is used as a positive control for the HBs probe (*lane 9). Mouse Cyclophilin A (protecting 105 bp) was used as a loading control. 1; marker, 2,3; yeast RNA controls minus/plus RNase treatment, 4; HR2-L4 brain, 5; HR2-L6 brain, 6; HR2-L3 brain, 7; HR2-L5 brain, 8; wild type brain. (C) RNase protection assay with probe HC in adult HR2-L3 transgenic organs showing relative abundance of spliced and unspliced human CGI-2 transcripts. The probe protects 121 bp spliced AIR transcript in lanes 5–8 (the band is weak in lane 5), and a 260 bp unspliced AIR transcript in lanes 5–8 (the band is weak in lane 5). Mouse 18S probe (protecting 80 bp) was used as a loading control. 1; marker, 2; HC probe, 3; yeast controls minus RNase, 4; liver, 5; lung, 6; brain, 7; heart, 8; testis, 9; kidney, 10; wild type testes control, 11; wild type kidney control. (D) Identification of human AIR transcriptional start sites in HR2-S2 transgenic mice by 5′RACE with gene specific primers 2f and 4f. Lane 1; minus phosphotase control, lane 2; plus phosphotase, lane 3; no template. T1, T2; transcription start site 1 and 2. (E) T1 and T2 start sites in human CGI-2 transgenic mice. nd; not done.