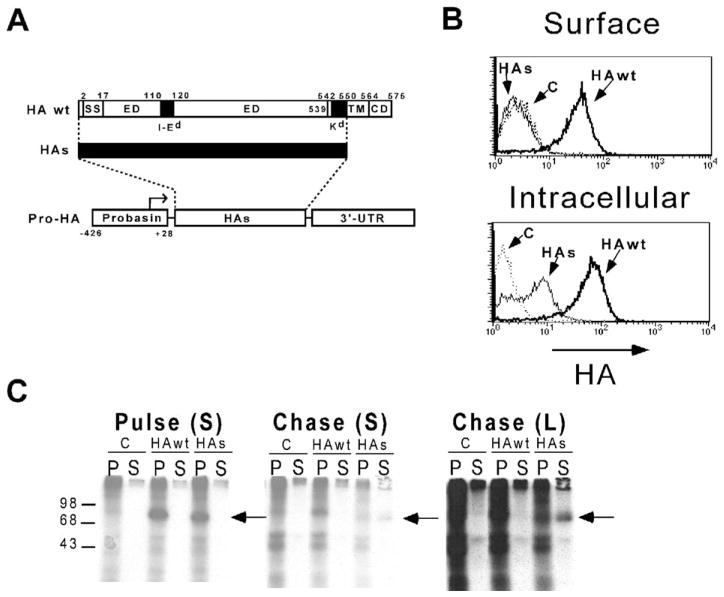
Figure 1. The Pro-HA expression construct.
A: The Pro-HA transgene, with the structure of the secreted HA (HAs) protein shown above. A map of the HA wt protein shows the locations of the ER signal sequence (SS, amino acids 2–17), the I-Ed epitope (amino acids 110–120, located within the extracellular domain [ED]), the Kd epitope (amino acids 542–550, located within the trans-membrane domain [TM, amino acids 539–564]), and the cytoplasmic domain (CD). The structure of the HAs protein is shown by a thick black line designating the portion of the wild-type protein that has been retained.
B: Subcellular localization of HA wt and HAs proteins. FACS histogram plots of P815 cells infected with the indicated recombinant vaccinia (or not infected as a background staining control [C]) and stained for HA either directly (Surface) or after membrane fixation and permeabilization (Intracellular).
C: Immunoprecipitation of HA wt and HAs proteins. COS-7 cells were transiently transfected with expression plasmids encoding the HA wt or HAs constructs, or the empty expression plasmid (C). Forty-eight hours later, they were labeled with [35S]methionine for 1 hr (pulse) and then cultured an additional 3 hr with an excess of cold methionine (chase). Following both the pulse and chase periods, cell pellets (P) as well as media supernatants (S) were harvested for immunoprecipitation using the anti-HA mAb H-18. Samples were analyzed by SDS-PAGE, with the location of molecular weight standards shown on the left, and the location of the HA proteins marked by an arrow to the right of each autoradiogram. Both short (S) as well as long (L) time exposures of the chase autoradiogram are shown.