Figure 1.
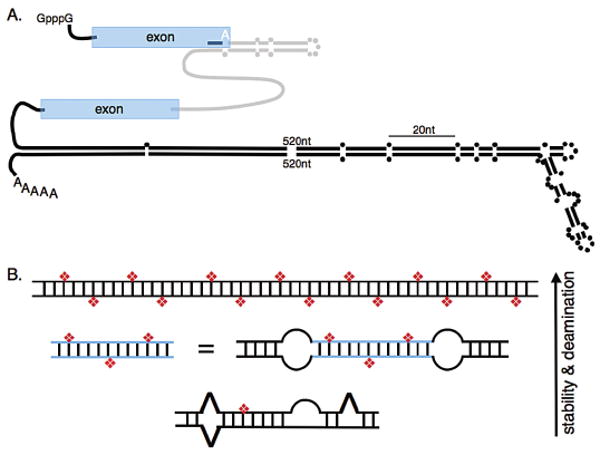
(A) The illustration depicts a pre-mRNA that contains examples of structures that are selectively (upper) and nonselectively (lower) edited. Coding exons are depicted in blue, an intron in gray, and 5′ and 3′ UTRs as black lines. Parallel lines represent base-paired regions with dots representing unpaired nucleotides. The selectively edited hairpin involves pairing between exon and intron sequences and is patterned after the R/G editing site, shown as a white A, found in certain mammalian glutamate receptor pre-mRNAs; the hairpin contains 28 bps, a loop, and mismatches as indicated (Aruscavage and Bass 2000). The nonselectively edited structure is patterned after the 3′UTR of the C. elegans gene, C35E7.6 (Morse et al. 2002). The edited structures are scaled relative to their actual lengths (20 nucleotides indicated), but 520 nucleotides of each strand of the 3′UTR structure were omitted as indicated. (B) The cartoon illustrates that the number of adenosines deaminated by an ADAR at reaction completion, or the selectivity of the enzyme, increases with the thermodynamic stability of the RNA structure. The top structure represents a long, completely base-paired dsRNA ≥ 50 bp which is deaminated nonselectively, showing 50–60% of its adenosines converted to inosines (red diamonds) at reaction completion. As indicated, structures that are less stable because they are shorter, or interrupted by mismatches, bulges, or loops, contain fewer inosines at the end of the reaction. Blue lines represent a specific sequence that exists as a separate molecule or between two internal loops of a longer structure.
