Abstract
Flexible ligands pose challenges to standard structure-activity studies since they frequently reorganize their conformations upon protein binding and catalysis. Here, we demonstrate the utility of side chain 13C relaxation dispersion measurements to identify and quantify the conformational dynamics that drive this reorganization. The dispersion measurements probe methylene 13CH2 and methyl 13CH3 groups; the latter are highly prevalent side chain moieties in known drugs. Combining these side chain studies with existing backbone dispersion studies enables a comprehensive investigation of µs–ms conformational dynamics related to binding and catalysis. We perform these measurements at natural 13C abundance, in congruence with common pharmaceutical research settings. We illustrate these methods through a study of the interaction of a phosphopeptide ligand with the peptidyl-prolyl isomerase, Pin1. The results illuminate the side-chain moieties that undergo conformational readjustments upon complex formation. In particular, we find evidence that multiple exchange processes influence the side chain dispersion profiles. Collectively, our studies illustrate how side-chain relaxation dispersion can shed light on ligand conformational transitions required for activity, and thereby suggest strategies for its optimization.
Keywords: Dynamics, Ligand, Relaxation dispersion, Side-chain, Drug-design
Introduction
In drug discovery, traditional structure-activity-relationship (SAR) investigations search for correlations between ligand structure and biological activity to drive ligand optimization. But such investigations can become complicated if the ligand can adopt more than one structure—that is, when the ligand is flexible. In such cases, the relationship between structure and activity can become ambiguous.
This ambiguity is a genuine concern in drug design: one often encounters flexible ligands in the course of lead generation and iterative optimization. Such ligands may be chemical probes used to extract functional information, or lead compounds under the process of optimization. Flexible ligands are also prominent in current drugs. Examples include peptido-mimetic compounds, such as Telaprevir, a current Phase III clinical candidate for HCV protease (Perni et al. 2007), as well as HIV-1 protease inhibitors (King et al. 2004). These ligands retain a number of rotatable bonds, whose torsion angle mobility allows for conformational readjustments upon protein binding. Indeed, an analysis of 150 crystal structures of flexible drug-like molecules bound to their protein targets by Perola and Charifson show that the bound ligand conformations often do not correspond to the calculated conformational energy minima of the free ligand (Perola and Charifson 2004). This means flexible ligands frequently undergo conformational reorganization upon binding. Such re-organization can lead to confounding or non-intuitive SAR.
To optimize the binding properties of flexible ligands, it would be useful to have methods that can describe in a site-specific manner where and how the ligand reorganizes upon complex formation. Such methods could illuminate which degrees of freedom come into play, what their relative amplitudes of motion are, and how they transform the free ligand conformational ensemble into the presumably narrower bound ligand ensemble. The resulting information could help estimate the reduction in the accessible ligand conformational space caused by binding, and provide insight into the ligand conformational entropy changes.
Accordingly, we have approached the problem of characterizing ligand conformational reorganization by exploiting the well-known power of NMR nuclear spin relaxation measurements to profile molecular dynamics. In particular, we have used 13C relaxation measurements to profile the internal flexibility of ligands in their free versus protein-complexed states. 13C relaxation parameters are appealing because they can be interpreted in a site-specific manner, and carbons are prevalent in drug-like molecules. Previously, we demonstrated methods to probe faster ps-ns motions of aromatic 13C methine (CH) nuclei, using natural abundance exchange-transferred cross-correlated relaxation (Peng 2003). More recently, we demonstrated the utility of natural abundance 13C Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion measurements to probe the slower µs-ms exchange-dynamics of methine (CH) 13Cα nuclei in peptide ligands upon protein binding (Zintsmaster et al. 2008).
Here, we look beyond the methine CH moiety to the methyl CH3 and methylene CH2 moieties. A retrospective study of the comprehensive medicinal chemistry (CMC) database by Bemis and Murcko show that CH3 moieties are among the most common side chains in drug-like molecules (Bemis and Murcko 1999). Methylene CH2 groups may be part of linkers or non-aromatic ring structures (Bemis and Murcko 1996). The prevalence of these moieties makes them worthy probes of ligand flexibility for molecules of pharmaceutical interest.
We demonstrate here the ability for natural abundance 13C dispersion measurements of side-chain CH3 and CH2 groups to illuminate the µs-ms conformational transitions required of the ligand for biological activity. This entails 13C relaxation dispersion studies of the interaction between the 18.4 kDa peptidyl-prolyl isomerase, Pin1, and a 10-residue phosphopeptide ligand that contains the target sequence from the Pin1 substrate, Cdc25 phosphatase. As with our previous studies, we work at natural 13C abundance (> 1%). While the dispersion methods are, of course, applicable to isotope-enriched ligands, such ligands are uncommon in pharmaceutical research settings, due to considerations of cost and synthetic complexity. Hence, it is important to demonstrate the feasibility of these dispersion experiments at 13C natural abundance. Historically, natural abundance measurements required ultra-high ligand concentrations (>10 mM). Fortunately, the availability of high magnetic fields and cryogenically-cooled probes have reduced these requirements to 1–2 mM. A notable side-benefit of natural abundance 13C is that homonuclear 13C–13C dipolar and scalar couplings, which can complicate relaxation measurements, are effectively eliminated.
Under ideal conditions, 13C dispersion experiments can provide the 13C chemical shift changes associated with complex formation. 13C chemical shifts are widely recognized as probes of local conformation (Cornilescu et al. 1999; de Dios et al. 1993; Spera and Bax 1991; Wishart and Sykes 1994; Xu and Case 2002) and there is now intense interest in chemical shifts as the principal drivers of overall protein conformation (Cavalli et al. 2007; Shen et al. 2008) and conformational fluctuations (Berjanskii and Wishart 2006). In this context, the 13CH3 and 13CH2 dispersion studies disclosed here shed light on side-chain moieties that must undergo conformational readjustments upon complex formation. Our broader aim is to combine the ligand dynamics perspective gained from ligand 13C relaxation measurements (for both backbone and side chain), with the protein dynamics perspective gained from more widespread protein studies. Combining the ligand and protein perspectives should provide a more comprehensive database for learning how to cope with non-intuitive SAR caused by ligand and/or protein flexibility.
Our report proceeds as follows. After a description of material and methods, we describe the Pin1/ligand system. We then present the results of the relaxation dispersion measurements for both 13CH3 and 13CH2 groups at two static field strengths and two temperatures. We discuss the extracted exchange parameters, and what they imply about complex formation. This includes follow-up studies involving a truncated construct of Pin1. We conclude with comments on the significance of these results in terms of a methodology for describing ligand conformational reorganization.
Materials and methods
Samples
NMR samples consisted of a 10-residue phosphopeptide ligand in the absence or presence of U-15N, 70%-2D human Pin1. The phosphopeptide, E1-Q2-P3-L4-pT5-P6-V7-T8-D9-L10 (purchased from QCB) contains the Pin1 pT-P target sequence from the mitotic phosphatase, Cdc25, a well-established Pin1 target (Crenshaw et al. 1998; Zhou et al. 2000).
U- 15N and 70% 2D Human Pin1 (1–163) and the truncated construct, Pin1-PPIase (40–163) were overexpressed in E coli strain BL21 (DE3) and purified chromatographically from the bacterial lysate. The truncated construct omits the first 39 residues corresponding to the WW domain (vide infra). U- 15N and 70% 2D enrichment was achieved by growing the cells in M9 media (90% D2O) containing 15NH4Cl as the sole nitrogen source. Details have been given elsewhere (Namanja et al. 2007). The 70% perdeuteration of the Pin1 aliphatic carbons reduces intermolecular proton dipolar relaxation losses that would otherwise compromise sensitivity, and further promotes selective ligand detection.
Samples containing Pin1 and the Cdc25 ligand consisted of 50 µM Pin1 and 2 mM Cdc25. Samples of the isolated ligand consisted of just 2 mM Cdc25. For all samples, the buffer conditions were 30 mM deutero-imidazole pH 6.6, 30 mM sodium chloride, 0.03% sodium azide, 1 mM deutero-dithiothreitol and 10% D2O.
Correct folding of Pin1 was assessed from the 15N–1H chemical shift dispersion in 2-D 15N-1H TROSY-HSQC spectra. Pin1 isomerase activity was confirmed from 2-D 1H-1H EXSY exchange cross-peaks between ligand 1H resonances corresponding to the trans and cis isomers. Integration of the 1-D 1H resonances of the trans and cis resonances at 295 K indicated ~ 10% cis population.
NMR experiments
We recorded all spectra on Bruker Avance systems (16.4 T and 18.8 T) equipped with TCI cryoprobes. We used established proton-detected pulse schemes to measure 13C relaxation dispersion for both methyl 13CH3 and methylene 13CH2 carbons. For the methyl 13CH3 experiments, we used the sequence by Lundström et al. (Lundström et al. 2007). For the 13CH2 experiments, we used the sequence by Mulder et al. (Mulder et al. 2001b). Both experiments use the relaxation-compensated Carr-Purcell-Meiboom-Gill (CPMG) spin-locking (Carr and Purcell 1954; Meiboom and Gill 1958), originally developed by Loria et al. (Loria et al. 1999). The relaxation-compensated schemes remove the dependence of the transverse relaxation rate on the CPMG inter-pulse delay, tcp, caused by oscillation between in-phase and antiphase coherences. The compensation thus assures that any variations in transverse relaxation rates with tcp reflect bona fide chemical exchange.
The dispersion experiments produce 2-D 13C–1H spectra in which the cross-peak intensity of each 13C nucleus is modulated by exp(−R2effTRELAX). R2eff is a rate constant that describes the effective transverse relaxation during a CPMG spin-lock of fixed length, TRELAX. For the dispersion studies, the key experimental parameter is the effective frequency of the CPMG spin-lock, νeff = 1/2tcp. The dispersion studies consist of measuring a series of 2-D spectra that correspond to increasing νeff (decreasing tcp). Analyses of the cross-peak intensities over the series produces for each 13C a R2eff versus νeff profile. Those carbons with R2eff versus νeff profiles displaying dispersion—a frequency dependence—are sites of chemical exchange, and are subjected to subsequent model-fitting for the extraction of site-specific exchange dynamics.
Generally, our dispersion studies included measurements at two static fields: 16.4T (700 MHz 1H Larmor frequency) and 18.8T (800 MHz 1H Larmor frequency). 13C hard pulses were applied at 17.86 kHz. The CPMG refocusing 180°s were applied at a lower strength of 6.7 kHz. In each dispersion series, we recorded duplicates of the reference spectra (spectra in which the CPMG is omitted) for subsequent uncertainty estimates.
13CH3 methyl dispersion experiments were acquired with the 13C carrier centered at 17.4 ppm and a 13C sweep-width of 18.2 ppm. Unless the temperature is specified explicitly below, the same parameters were used at both 278 and 295 K. We generally acquired 32 scans per increment, and a total of 26 complex points, resulting in 1 h per 2-D spectrum. For Cdc25 in the presence of full-length Pin1, we used TRELAX = 88.1 ms, and the νeff = 1/2tcp values were as follows: 46, 92, 139, 187, 235, 284, 334, 385, 436, 487, and 540 Hz. For Cdc25 in the presence of Pin1-PPIase, we used a longer TRELAX = 120 ms, with νeff = 34, 67, 102, 136, 171, 206, 242, 278, 314, 351, 426, 502, and 540 Hz. For the isolated ligand we took a smaller range of νeff samples, after observing no significant changes between the lowest and highest νeff values. At 278 K, 16.4 T, TRELAX = 120 ms, and νeff values included 34, 67, 102, 136, 351, 426, and 540 Hz, while at 295 K, 16.4T, TRELAX = 88.1 ms, and νeff values were 46, 92, 187, and 540 Hz.
For the 13CH2 methylene dispersion experiments, the 13C carrier was centered at 48 ppm. For Cdc25 in the presence of full-length Pin1, we used TRELAX = 40.0 ms, with νeff = 1/2tcp values as follows: 50, 102, 153, 206, 260, 314, 369, 426, 483, and 540 Hz. At 16.4 T, 295 K we took 384 scans per increment (5.6 h per 2-D); in all other series we acquired 256 scans per increment (3.7 h per 2-D). In all cases, we collected a total of 14 complex points over a 13C sweep-width of 56.8 ppm. For Cdc25 in the presence of Pin1-PPIase, we used a longer TRELAX = 64 ms, with νeff = 31, 63, 95, 127, 193, 328, 468, and 540 Hz. For the isolated Cdc25 ligand, we saw no significant intensity changes between the highest and lowest νeff values, and so used a smaller sampling of νeff values. At 278 K, 16.4T, we used TRELAX = 40 ms with νeff = 50, 206, 314, 426, and 540 Hz. At 295 K, 16.4T, we used TRELAX = 64 ms, with νeff = 31, 63, 95, 127, 193, 328, 468, and 540 Hz.
We also performed 1H–1H EXSY spectra to verify Pin1 isomerase activity on the Cdc25 phosphopeptide substrate. The exchange mixing time employed a continuous-wave spin-lock (as in ROESY), bracketed by 4 ms 1H adiabatic rotations (Mulder et al. 1998). The spin-lock field strength was 2.0 kHz, and was centered on the methyl protons of pT5. These protons showed resolved 1H resonances corresponding to the major trans and minor cis populations in 1-D spectra. Exchange mixing times (i.e. the entire time interval between the t1 and t2 chemical-shift evolution periods), included: 22, 31, 41, 61 (2×), 72 (2×), 81 (2×), 101, 121 (2×), 171, and 221 ms. Selective gradient-echo sandwiches (p90—gradient—sel-p180—gradient) prior to t1 and t2 allowed narrower spectral widths of 3.57 ppm in both dimension. The sel-p180 was an 8 ms REBURP pulse with a peak amplitude of 500 Hz (Geen and Freeman 1991).
Data reduction
All spectra were processed using Bruker Topspin 1.3 software. Subsequent measurement of cross-peak intensities, extraction of relaxation rates, and fitting of exchange parameters used in-house C-programs.
For resolved 13C–1H cross-peaks, we obtained R2eff versus νeff = 1/2tcp profiles from the relation R2eff(1/2tcp) = −1/TRELAX × ln{I(1/2tcp)/Iref} (Mulder et al. 2001a). I(1/2tcp) and Iref are the cross-peak intensities in the presence and absence of the CPMG spin-lock. Uncertainties in the rate constants were estimated by Monte Carlo simulations, based on spectral duplicates.
We fit the resulting R2eff versus νeff profiles to the general two-state exchange model, using the Carver-Richards expressions (Carver and Richards 1972; Millet et al. 2000). The Carver-Richards expressions apply to all time scales of exchange, and consist of four adjustable parameters: the net exchange rate constant, kex; the fractional population of the minor species, pB; the chemical shift difference between the two putative states, Δppm; and R2,0, the nominal non-exchange contributions to R2eff, including relaxation caused by heteronuclear dipole-dipole interactions and anisotropic chemical shielding. To find the optimal parameters, we used in-house software to minimize the χ2 error via a grid-search algorithm (Bevington 1969). For global fits, we kept kex and pB the same for all 13C nuclei, while letting Δppm and R2,0 vary in a site-specific manner. Uncertainties in the resulting parameters were estimated by jack-knife simulations.
We obtained independent estimates of the exchange rate constants for cis-trans isomerization using aforementioned series of 2-D 1H–1H EXSY spectra. We fit the ratio of the exchange cross-peak intensity over the diagonal cross-peak intensity as a function of mixing time to the two-state EXSY expressions of Ernst et al. (Ernst et al. 1987) This yielded rate constants ktc (trans to cis) and kct (cis to trans). From these rate constants, we obtained the net cis-trans isomerization rate constant kex,EXSY = ktc + kct, which is the rate constant appropriate for comparison with kex extracted from 13C relaxation dispersion. Uncertainties in the rate constant were estimated via Monte Carlo simulations based on spectral duplicates.
Results and discussion
Our 13C dispersion experiments focus on the interaction of a ten-residue phosphopeptide, E1-Q2-P3-L4-pT5-P6-V7-T8-D9-L10, with the protein, Pin1. Pin1 is an 18.4 kDa peptidyl-prolyl isomerase that accelerates the cis-trans isomerization of phosphorylated-Threonine/Serine-Proline (pT/S-P) motifs in other cell signaling proteins (Lu et al. 2007). Pin1 has a modular design, consisting of a catalytic peptidyl-prolyl isomerase (PPIase) domain that is flexibly linked to a smaller WW domain. The PPIase domain is solely responsible for isomerase activity, while the WW domain functions as a docking module specific for pT/S-P motifs.
Our phosphopeptide ligand represents a pT-P motif from the mitotic phosphatase Cdc25, a well-known Pin1 target (Crenshaw et al. 1998; Zhou et al. 2000). Its Pin1 binding affinity is characterized by KD = 8 ± 2 µM at 295 K (imidazole buffer pH 6.6). The KD was determined by fitting Pin1 15N–1H chemical shift perturbations versus ligand concentration in the course of a ligand titration (Namanja 2009). Additionally, previous NMR studies of this phosphopeptide confirm the interaction of the pT5-P6 linkage with Pin1 (Namanja et al. 2007; Wintjens et al. 2001). Moreover, our own 1H–1H EXSY experiments show explicit cis-trans isomerization (vide infra). Thus, the Cdc25 phosphopeptide/Pin1 system is an illuminating choice for our dispersion studies, since we know at the outset that their mutual interaction perturbs the ligand conformational ensemble.
We measured 13C relaxation dispersion profiles, at natural 13C abundance ( ~ 1%), for methyl and methylene carbons of the Cdc25 ligand, using established 2-D 13C–1H proton-detected pulse schemes. We examined the ligand in its isolated (free) state, and in the presence of Pin1. Samples containing both protein and ligand had a 40-fold molar excess of Cdc25 ligand (2 mM) over protein (50 µM). Since the total ligand concentration is much greater than KD (8 ± 2 µM at 295 K, imidazole buffer, pH 6.6) we can safely neglect the free Pin1 concentration. The 40-fold ligand molar excess of ligand also serves to enhance the selective observation of ligand 13C resonances.
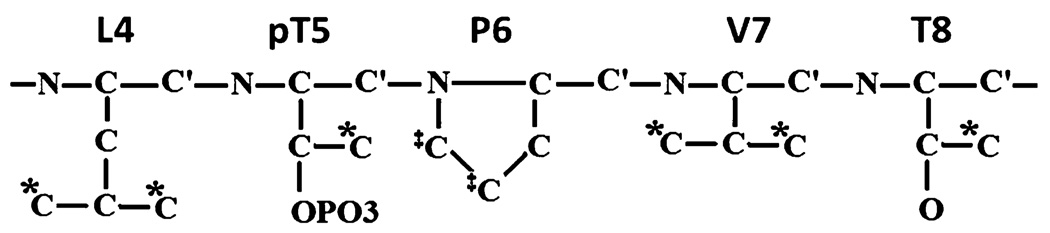
Figure 1 schematizes the five core residues of the Cdc25 ligand. The asterisks and double-daggers highlight the side-chain methyl and methylene carbons, respectively. Critically, methylene and methyl carbons bracket the isomerization epicenter—the pT5-P6 peptide linkage. Thus, it is reasonable to investigate the potential of these side-chain carbons as probes of µs-ms exchange dynamics via 13C relaxation dispersion measurements.
Fig. 1.
Schematic of the five core residues of Cdc25 phosphopeptide ligand, EQPLpTPVDL. Asterisks and daggers denote the methyl and methylene carbons highlighted in the 13C dispersion studies
Methyl dispersion measurements
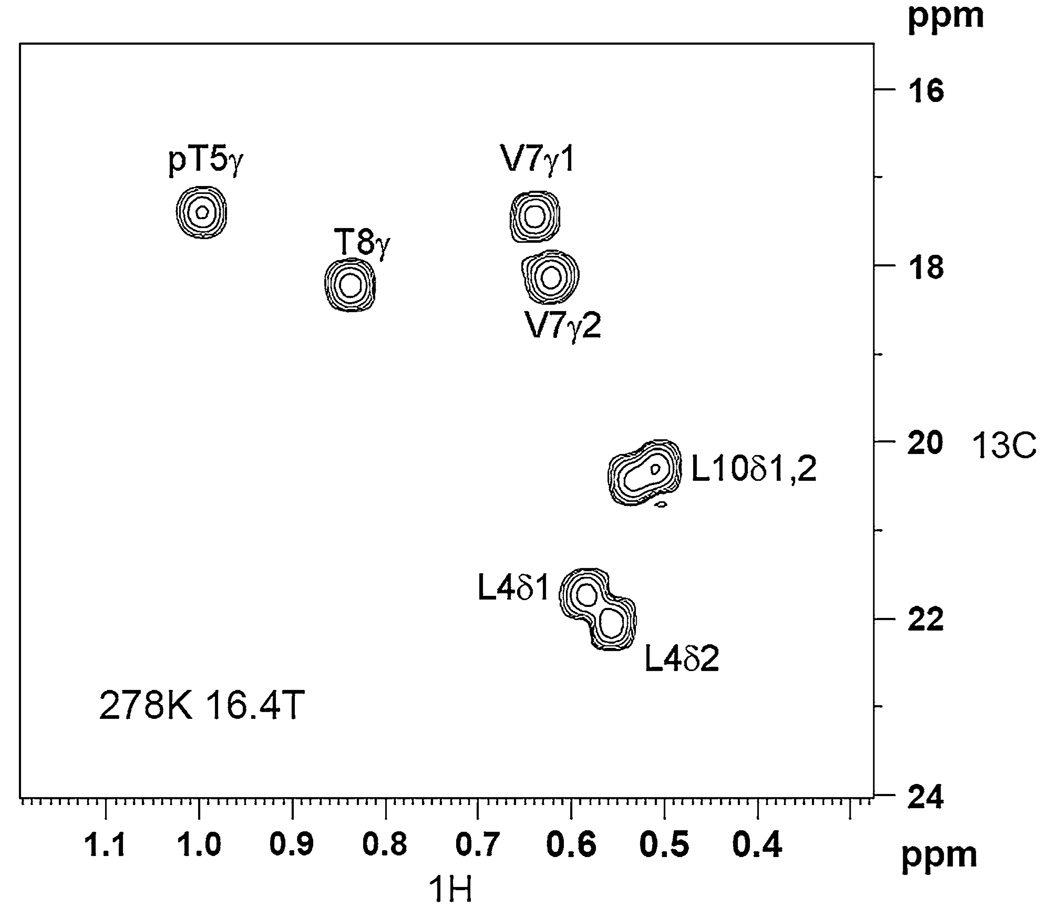
We used the 2-D 13C-1H relaxation dispersion scheme of Lundström et al. (Lundström et al. 2007) for our studies of the Cdc25 ligand methyl carbons. Cdc25 ligand has eight methyl groups. Six of those eight give sufficiently resolved 13C–1H cross-peaks in the corresponding dispersion spectra sufficient for analyses: L4-Cδ1/δ2, T5-Cγ, V7-Cγ1/γ2, and T8-Cγ. The Cδ methyl cross-peaks of L10 suffered severe overlap and were excluded from further analyses. Figure 2 shows an example spectrum of the Cdc25 methyls (in the presence of Pin1) at 16.4T (700 MHz 1H Larmor frequency), with tcp set to its longest value of 5.46 ms. The measurement time was 1 hr. We do not yet have stereospecific methyl carbon assignments, and so δ1/δ2 and γ1/γ2 simply indicate 13C–1H cross-peaks that are downfield versus upfield along the 1H chemical shift axis. For both the isolated ligand and the ligand in the presence of Pin1, we observe only one 13C–1H cross-peak per methyl group.
Fig. 2.
Example spectrum from the natural abundance 13CH3 methyl dispersion measurements of the Cdc25 ligand (2 mM), in the presence of 50 µM U-15N/70%2D Pin1. The spectrum is recorded at 278 K, 16.4 T. The 2-D represents the lowest CPMG frequency νeff = 46 Hz. The total CPMG duration was 88 ms
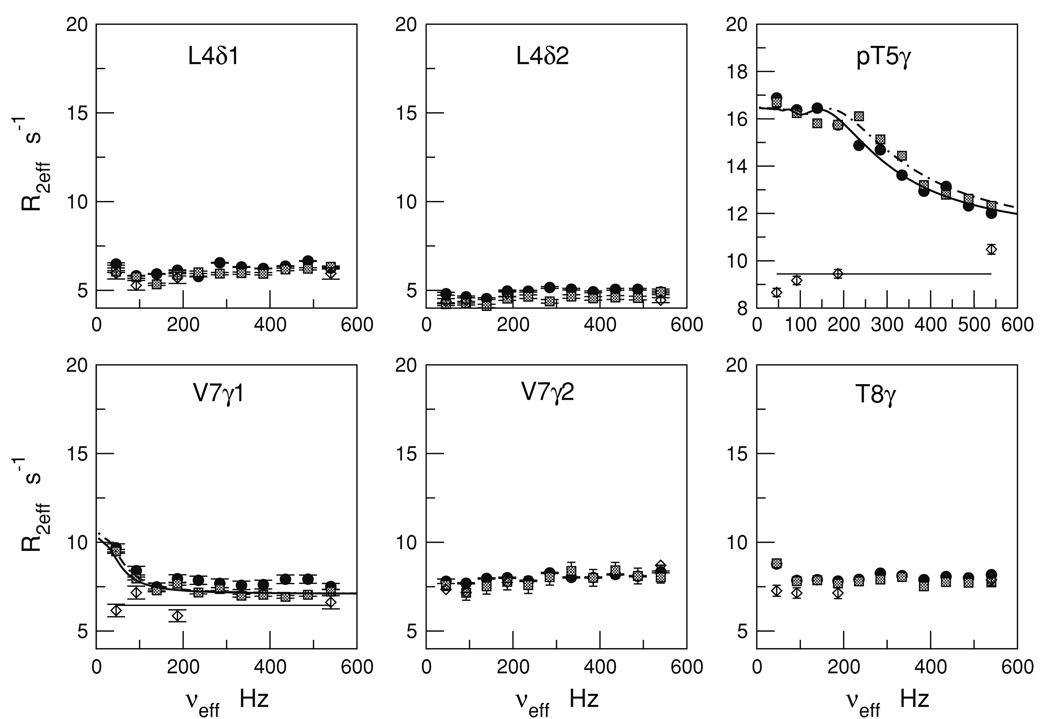
Figure 3 summarizes the R2eff versus νeff profiles for the six well-resolved methyl cross-peaks of the Cdc25 ligand at 278 K. The profiles include dispersion measurements at both 16.4 T (filled circles), and at 18.8 T (hatched squares).
Fig. 3.
13CH3 CPMG relaxation dispersion profiles for the Cdc25 ligand methyls with and without Pin1 at 278 K. The profiles are R2eff versus νeff = 1/2tcp where, tcp is the delay between consecutive 180 pulses in the CPMG spin-lock. Ligand R2eff in the presence of Pin1 are indicated by filled circles (16.4 T) and hatched squares (18.8 T). Free ligand R2eff (Pin1 absent) at 16.4 T are indicated by the diamonds. The curves are fits to the two-state exchange model using the Carver-Richards expressions (see text). The horizontal lines are the average R2eff values for the free ligand
The isolated ligand shows no significant dispersion—the R2,eff versus νeff profiles are essentially flat for all methyls. These individual and average values are indicated by the open diamonds and solid horizontal lines, respectively, in Fig. 3. The average and rmsd values of R2eff at 278 K are as follows: L4-Cδ1 (5.7 ± 0.2 s−1), L4-Cδ2 (4.5 ± 0.1 s−1), T5-Cγ (9.4 ± 0.5 −1), V7-Cγ1 (6.5 ± 0.4 −1), V7-Cγ2 (7.7 ± 0.4 −1), and T8-Cγ (7.4 ± 0.2 −1). In the absence of exchange broadening, R2eff should reflect the ps-ns reorientational dynamics of the methyl 13C–1H bond vectors. Hence, the variations in the free ligand R2eff most likely reflect the different number of torsion angles separating them from the ligand backbone.
The situation changes in the presence of Pin1. Most methyl carbons remain unaffected. However, pT5-Cγ and V7-Cγ1 show strong dispersion responses. These dispersion responses occur only upon introduction of Pin1, and are thus a direct consequence of Pin1/Cdc25 ligand interaction. More precisely, interaction with Pin1 at 278 K results in a ligand exchange process that modulates the isotropic 13C chemical shifts of both pT5-Cγ and V7-Cγ1.
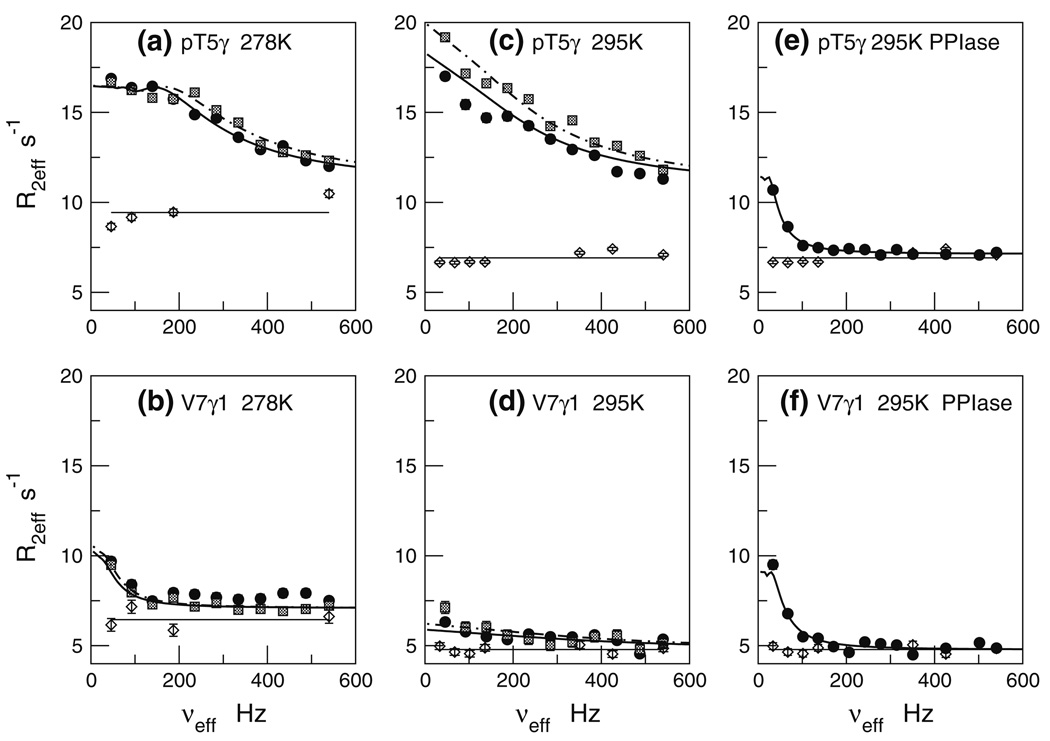
We repeated the methyl dispersion experiments at higher temperature of 295 K. As before, pT5-Cγ and V7-Cγ1 show clear dispersion profiles, while the other methyls are unresponsive. Figure 4 juxtaposes the R2eff vs νeff profiles at 278 K (panels a, b) versus 295 K (panels c, d), respectively. For pT5-Cγ, we see somewhat larger R2eff values at 295 K, suggestive of faster chemical exchange at 295 K versus 278 K.
Fig. 4.
13CH3 CPMG relaxation dispersion profiles for pT5-Cγ and V7-Cγ1 under various conditions. Panels a, b: ligand +/− full-length Pin1 at 278 K; Panels c, d: ligand +/− full-length Pin1 at 295 K; Panels e, f: ligand +/− Pin1-PPIase at 295 K. Symbols, curves, and horizontal lines have the same meaning as in Fig. 3
The selective dispersion pT5-Cγ and V7-Cγ1 raises the question: what distinguishes these methyls from other ligand methyls in the context of the Pin1/ligand interaction? To address this question, we can turn to the solution structure of the Cdc25 phosphopeptide ligand complexed with the Pin1-WW domain (Wintjens et al. 2001; PDB ID code 1I8G). This structure shows that pT5-Cγ and V7-Cγ1 are intimately involved in the protein-ligand contact interface, while the other methyl groups are remote. To the extent that these same interactions are preserved when Cdc25 is complexed with the full-length Pin1, this structure provides a basis for understanding the selective dispersion responses of pT5-Cγ and V7-Cγ1.
Methylene dispersion measurements
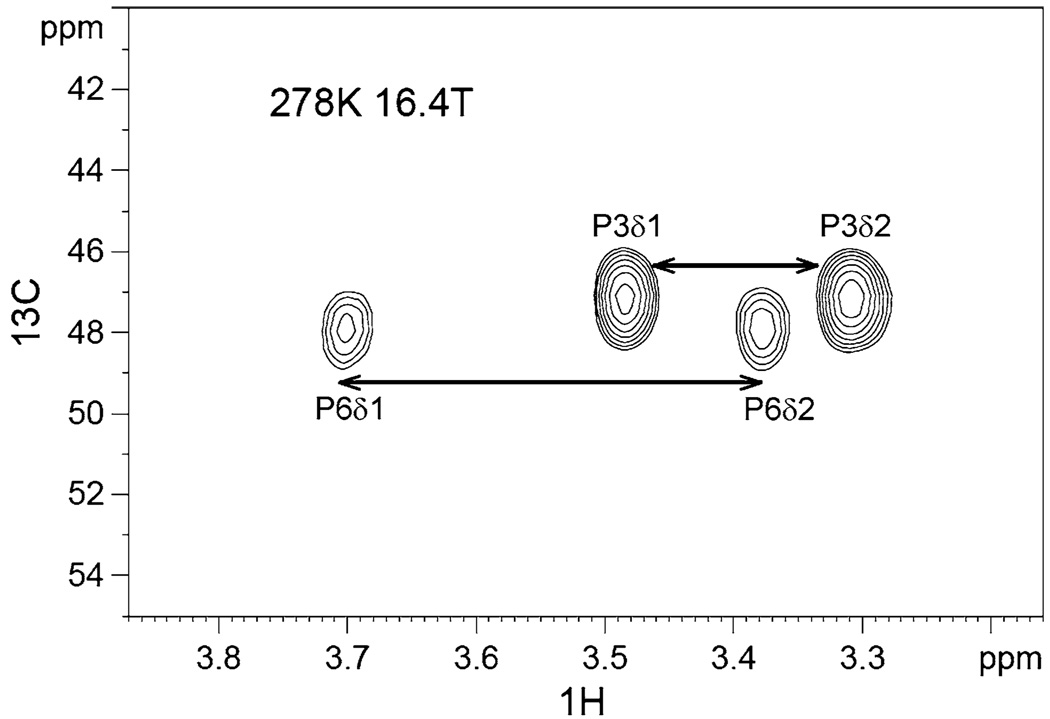
For the methylene dispersion measurements, we used the 2-D relaxation dispersion scheme of Mulder et al. which was originally demonstrated on protein 15NH2 groups (Mulder et al. 2001b). The 13C methylene chemical shifts span a considerable range of ~ 25 ppm. If one is to perform CPMG spin-locking for all methylenes over this range and simultaneously avoid off-resonance artifacts, then the prudent course would be to carry out multiple CPMG dispersion studies with the CPMG spin-lock carrier centered at multiple positions. However, for the Cdc25 ligand, the 13C methylene experiments proved considerably less sensitive than the 13C methyl experiments. The lower sensitivity limited the time available for multiple CPMG studies. Thus, we focus here only on the proline methylene carbons, P3-Cδ and P6-Cδ, which have nearly the same 13C chemical shifts. Notably, P6-Cδ is a priori of interest since it is part of the signature pT-P motif that undergoes Pin1-catalyzed cis-trans isomerization. Figure 5 shows an example spectrum of P3-Cδ and P6-Cδ (in the presence of Pin1), with tcp set to its longest value of 4.96 ms.
Fig. 5.
Example spectrum from the natural abundance 13CH2 methylene dispersion measurements of the Cdc25 ligand (2 mM), in the presence of 50 µM U-15N/70%2D Pin1. The spectrum is recorded at 278 K, 16.4 T. The 2-D represents the lowest CPMG frequency νeff = 50 Hz. The total CPMG duration was 40 ms. P6-Cδ clearly suffers line-broadening relative to P3-Cδ
We note that the sensitivity of the methylene experiment is further compromised by the fact that the NMR signal from one methylene carbon is divided among two 13C–1H correlations. Thus, as a compensatory measure, we first added the two cross-peak integrals of each methylene prior to extraction of the R2,eff relaxation rate constants.
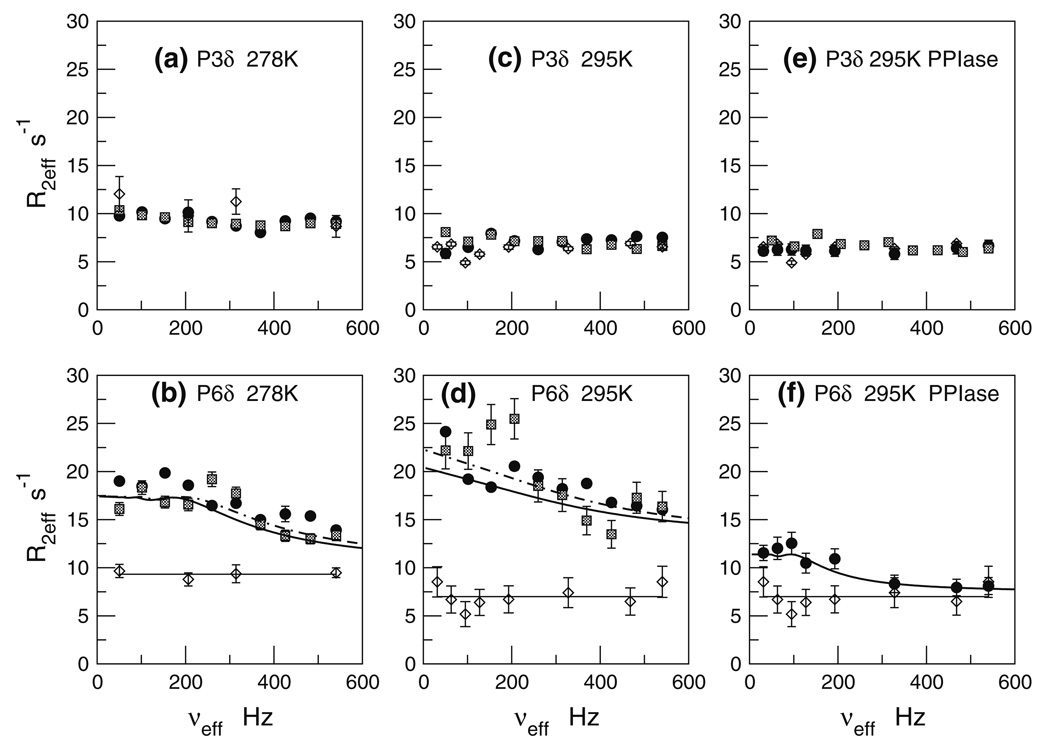
Figure 6a and b shows the resulting R2,eff versus νeff profiles at 278 K. The scatter and error bars reflect the lower sensitivity of the methylene experiments, relative to the methyl dispersion. For the isolated ligand, we observe no significant dispersion for either P3-Cδ or P6-Cδ. The R2,eff versus νeff profiles are essentially flat for both methylenes. The average and rmsd values for R2eff at 278 K are similar for both methylenes: P3-Cδ (10.4 ± 0.9 s−1) and P6-Cδ (9.3 ± 0.3 s−1).
Fig. 6.
13CH2 CPMG relaxation dispersion profiles for P3-Cδ and P6-Cδ under various conditions. Panels a, b: ligand +/− full-length Pin1 at 278 K; Panels c, d: ligand +/− full-length Pin1 at 295 K; Panels e, f: ligand +/− Pin1-PPIase at 295 K. Symbols, curves, and horizontal lines have the same meaning as in Fig. 3. P3-Cδ retains R2eff values of the free ligand under all conditions
Upon interaction with Pin1, there is a marked change: P6-Cδ shows clear dispersion. By contrast, P3-Cδ remains unaffected. Indeed, this differential behavior is foreshadowed by the differential line-broadening of P6 in Fig. 5.
We repeated the methylene dispersion experiments at the higher temperature of 295 K, and the results are in Fig. 6c and d. The selective dispersion response of P6-Cδ, while noisy, is nevertheless present. The larger scatter and error bars for the 18.8T R2eff values (hatched squares) are due mainly to poor solvent suppression. We note that the 295 K R2eff values (Fig. 6c, d) increase from those at 278 K (Fig. 6a, b). This R2eff increase supports the notion of slow exchange for P6-Cδ at 278 K, nudging towards faster exchange at 295 K. By contrast, the R2eff of the isolated ligand decreases relative to that at 278 K. Such decreases are of course expected if the free ligand R2eff lacks contributions from chemical exchange broadening. Then, R2eff reflects mainly ps-ns motions of the CH bond vector, and so the lower R2eff likely reflects primarily more rapid tumbling of the ligand at 295 versus 278 K.
The clear dispersion response of P6-Cδ, and the utter lack of response by P3-Cδ is consistent with P6-Cδ being part of the pT-P motif targeted by Pin1. Moreover, the solution structure of the Pin1-WW/Cdc25 ligand complex shows that P6-Cδ forms the central portion of the binding interface, while the P3-Cδ is comparatively remote (Wintjens et al. 2001). Thus, preferential chemical shift modulation of P6-Cδ over P3-Cδ is reasonable.
Extraction of exchange parameters
The above side chain R2eff vs νeff profiles show that the natural abundance 13CH3 and 13CH2 dispersion data are sensitive to the µs-ms dynamics associated with complex formation of a ligand. Specifically, in the presence of Pin1, the methyl carbons of the Cdc25 ligand T5-Cγ and V7-Cγ1 as well as its methylene carbon P6-Cδ, show temperature-sensitive dispersion profiles. By contrast, the isolated ligand shows no dispersion. Interaction of the Cdc25 ligand with Pin1 selectively modulates the chemical shifts of T5-Cγ, V7-Cγ1 and P6-Cδ. All of these carbons reside within or flank the pT5-P6 imide linkage, which undergoes Pin1-catalyzed cis-trans isomerization. Additionally, previous structural studies show that these carbons reside within side-chains that make contact with Pin1 (Wintjens et al. 2001). 13C chemical shifts are sensitive probes of local structure (torsion angles; Cavalli et al. 2007; Shen et al. 2008; Wishart and Sykes 1994; Xu and Case 2002); hence, the side-chain dispersion profiles reflect, at least in part, ligand conformational reorganization caused by complex formation with Pin1.
To extract quantitative exchange parameters, we fit the R2eff versus νeff dispersion profiles to model functions corresponding to the simplest scenario of two-state exchange. We applied the Carver-Richards model expressions, which contains four adjustable parameters: the net exchange rate constant kex, the minor state fractional population pB, the chemical shift difference between the two states Δppm, and finally the nominal non-exchange relaxation rate R2,0, which is independent of νeff = 1/2tcp.
For initial guidance, we turned to our recent dispersion measurements of the backbone methine 13Cα carbons, for the very same Pin1/Cdc25 interaction (Zintsmaster et al. 2008). In that previous study, Carver-Richards fits of the individual 13Cα methines (L4, pT5, P6, and P8) gave very similar exchange parameters. The similarity suggested the appropriateness of a global fit, in which the 13Cα nuclei share the same exchange rate constant kex and minor species fraction, pB, but vary site-specifically for the parameters Δppm and R2,0. Accordingly, after a global fit analysis, we find a global exchange rate constant kex = 477 ± 25 s−1 and a minor state fractional population of pB = 2.4 ± 0.2%. The results are listed in Table 1. Notably, the pB = 2.4 ± 0.1% coincides with bound ligand fraction, assuming Pin1-saturated conditions. As stated, this assumption is well-founded since our total ligand concentration (2 mM) is much greater than KD (8 ± 2 µM). It is therefore reasonable to identify the minor state sensed in the backbone dispersion studies as the Pin1-bound ligand. The strong dependence of 13Cα shifts on local torsion angles (φ, ψ, χ1) is well-established (Cornilescu et al. 1999; de Dios et al. 1993; Spera and Bax 1991; Vila and Scheraga 2008; Xu and Case 2002); hence, the methine dispersion likely reflects backbone torsion angle distortions of the ligand upon Pin1 complex formation.
Table 1.
Ligand backbone 13Cα global exchange parameters at 278 K
| Cα | kex (r/s) | pB | Δppm | R2,0 s−1 |
|---|---|---|---|---|
| L4 | 477 ± 25 | 0.024 ± 0.001 | 1.66 ± 0.04 | 9.5 ± 0.3 |
| pT5 | 477 ± 25 | 0.024 ± 0.001 | 0.52 ± 0.02 | 11.7 ± 0.1 |
| P6 | 477 ± 25 | 0.024 ± 0.001 | 0.66 ± 0.02 | 11.1 ± 0.1 |
| T8 | 477 ± 25 | 0.024 ± 0.001 | 0.67 ± 0.02 | 8.5 ± 0.1 |
We then used the backbone-derived kex and pB values as initial values for the side-chain 13CH3 and 13CH2 dispersion data. To our frustration, we found that enforcing one, but not both of these backbone 13Cα-derived global values produced satisfactory fits. Nevertheless, we reasoned that it was prudent to identify the minor state as the bound ligand fraction, just as in the backbone studies. Consequently, we set pB = 2.4% and proceeded with the parameter-fitting. The solid and dashed-dotted curves in Fig. 4 and Fig. 6 shows fits for pB = 2.4 ± 0.1%; and the results are listed in Table 2. They suggest that at 278 K, T5-Cγ, V7-Cγ1, and P6-Cδ undergo slow exchange on the 13C chemical-shift time scale, and transition to fast exchange at 295 K. Specifically, at 278 K, the side chain kex rate constants range from 200 to 300 s−1, while at 295 K they range from ~ 1,400 to ~ 2,500 s−1.
Table 2.
Ligand 13CH3 and 13CH2 exchange parameters with full-length Pin1 (Assumes pB = 0.024 ± 0.001)
| Carbon | T (K) | kex (r/s) | Δppm | R2,0 s−1 |
|---|---|---|---|---|
| pT5-Cγ, methyl | 278 | 230 ± 6 | 1.64 ± 0.30 | 11.0 ± 0.1 |
| V7-Cγ1, methyl | 278 | 200 ± 7 | 0.26 ± 0.01 | 7.1 ± 0.2 |
| P6-Cδ, methylene | 278 | 299 ± 15 | 1.93 ± 0.08 | 10.4 ± 0.4 |
| pT5-Cγ, methyl | 295 | 1,360 ± 246 | 0.67 ± 0.05 | 10.9 ± 0.5 |
| V7-Cγ1, methyl | 295 | 2,480 ± 570 | 0.31 ± 0.05 | 4.8 ± 0.2 |
| P6-Cδ, methylene | 295 | 2,080 ± 233 | 0.79 ± 0.05 | 13.1 ± 0.34 |
What exchange processes underlie the dispersion profiles? At least two processes must be considered: binding to Pin1, and cis-trans isomerization by Pin1. We estimated the cis-trans isomerzation rates from 2-D 1H–1H EXSY spectra of Pin1, focusing on the protons directly attached to pT5-Cγ. The EXSY exchange rate constants, kex,EXSY, are calculated from exchange cross-peaks that connect major (pT5-Hγtrans) and minor (pT5-Hγcis)) resonances of pT5 (spectra not shown). Hence, kex,EXSY is a more exclusive reporter of cis-trans isomerization. We found kex,EXSY = 5.5 ± 0.3 s−1 (278 K) to kex,EXSY = 8.8 ± 0.4 s−1 (295 K). These values are quite low compared to the 13C side-chain dispersion derived kex values in Table 2. Thus, the kex values from the side-chain 13CH3 and 13CH2 dispersion profiles, as well as the backbone, most likely reflect binding rather than cis-trans isomerization. This is reasonable, given that exchange rate constant for ligand binding, under protein saturated conditions, is kex,binding = konKD. Assuming the on-rate kon is diffusion-limited, then kon ≤ 1 × 108 M−1 s−1 (Davis et al. 1994). As stated, the Cdc25 ligand KD = 8 ± 2 µM, and so we would expect kex,binding to be ≤800 s−1. This estimate is in far better agreement with the side chain 13C dispersion derived kex values of 200–300 s−1 and the backbone kex values of 477 ± 25 s−1. At the same time we acknowledge the mismatch between these side chain and backbone kex values. The mismatch suggests that side chain and backbone experience different exchange processes.
Further interpretation of these results demands caution. As stated, Pin1 consists of two domains: a catalytic PPIase domain, and the smaller WW domain. Both domains can bind the pT-P motif. The distinction is that the PPIase domain is solely responsible for cis-trans isomerization. Thus, the 13C dispersion data could reflect interaction with either the PPIase or WW domain active sites. Moreover, the complex formation with the PPIase domain could involve conformational adjustments pertinent to both binding and cis-trans isomerization. These issues raise the possibility that the dispersion profiles in Fig. 4a–d and in Fig. 6a–d reflect more than one exchange process—that is, the exchange scheme may involve more than two chemical states.
Comparison with truncated Pin1 construct indicates multiple exchange processes
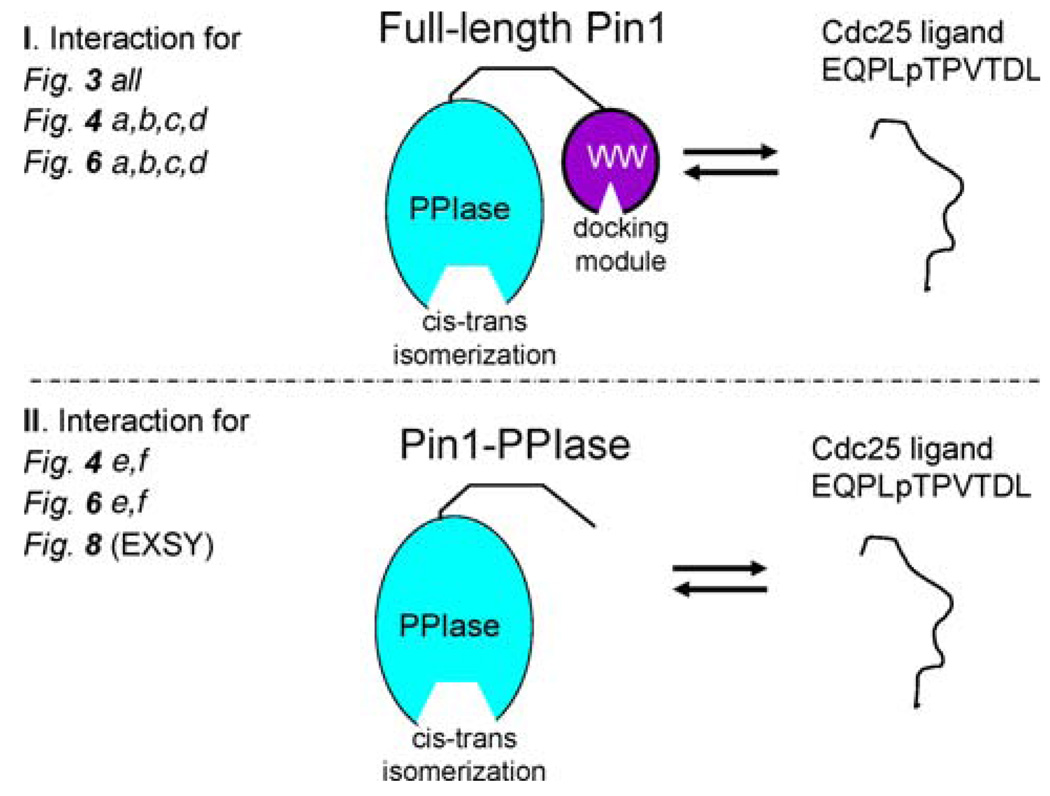
To shed light on the above possibility, we generated a truncated Pin1 construct that omits the Pin1-WW domain. We refer to this shortened construct as Pin1-PPIase. Pin1-PPIase is necessary and sufficient for in vitro isomerase activity (Labeikovsky et al. 2007; Verdecia et al. 2000). This construct removes the effects of WW domain binding; hence, the 13C dispersion profiles solely reflect interaction with the PPIase active site. Figure 7 schematizes this shorter construct, and contrasts it with the full-length situation above. Henceforth, Pin1-PPIase refers to the bottom panel of Fig. 7, while Pin1 (i.e., the full-length protein that has been discussed in the preceding sections above) refers to the top panel of Fig. 7.
Fig. 7.
Schematic of interactions described in this study. The top interaction (I): ligand +/− full-length Pin1. Full-length Pin1 has two domains, both of which can individually bind the ligand; the smaller WW domain (purple), which serves as a pure docking module, and the larger PPIase domain (aqua), which is solely responsible for cis-trans isomerization. The full-length Pin1 interaction is pertinent to the 13C dispersion studies in all of Fig. 3, Fig. 4a–d, and Fig. 6a–d. The bottom interaction (II): ligand +/− Pin1-PPIase. The shorter Pin1-PPIase construct omits the WW domain, thus simplifying the system to just the active site of the PPIase domain. The Pin1-PPIase interaction is pertinent to the 13C dispersion studies profiled in Fig. 4e and f and 6e, f, as well as the 2-D 1H–1H EXSY study of Fig. 8
We repeated the 13C methyl and methylene dispersion experiments for the Cdc25 ligand at 295 K, 16.4 T (700 MHz) in the presence of Pin1-PPIase. The resulting R2eff versus νeff dispersion are in Fig. 4e and f and Fig. 6f. Comparisons with Fig. 4c, d for the methyls, and Fig. 6d for the methylenes, show that the Pin1-PPIase profiles are markedly different from that seen with full-length Pin1. These differences are particularly evident for the methyls. First, the maximum R2eff value for pT5-Cγ in Pin1-PPIase (panel e) is much lower than that previously seen for full-length Pin1 (panel c). Second, the dispersion curve descends to its plateau at much lower frequency (νeff < 100 Hz) than in full-length Pin1, suggesting a significantly slower kex value. Third, the R2eff values at higher νeff (shorter tcp) are now same as that of the isolated (free) Cdc25 ligand. This last point is critical: recall the free ligand shows no significant dispersion. Hence, we are assured that the plateau R2eff values for pT5-Cγ and V7-Cγ1 in Fig. 4e and f are “exchange-free”. In the case of interactions with full-length Pin1, only V7-C γ1 at 295 K came close to such behavior.
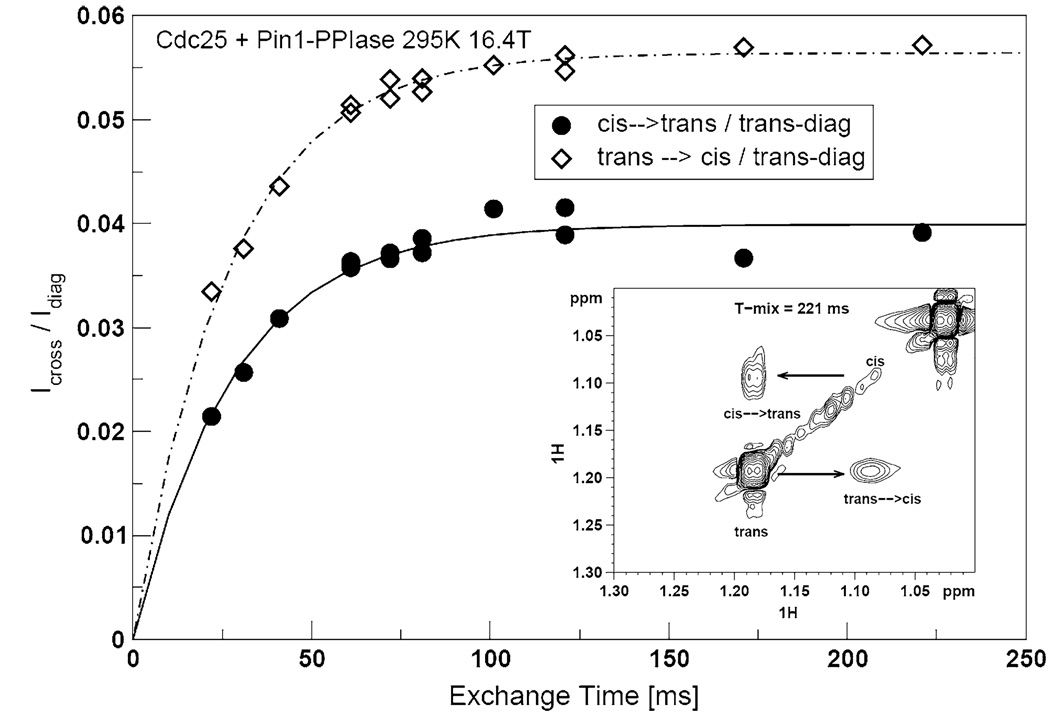
We then tried to fit the pT5-13Cγ and V7-13Cγ1 methyl dispersion profiles to the Carver-Richards two-state exchange model. The kex values, previously obtained from the studies with full-length Pin1 (Fig. 4c, d), were far too large to produce satisfactory fits. Consequently, we tried the cis-trans isomerization rate constant, kex,EXSY = 37 ± 1.1 s−1, obtained from the 2-D 1H–1H EXSY spectra of pT5-1Hγ, in the presence of the shorter Pin1-PPIase construct at 295 K. (Note that this EXSY experiment, performed on the truncated Pin1-PPIase construct, is distinct from that already described for full-length Pin1). Figure 8 shows an example of the EXSY spectra and corresponding build-up curves. We also used the free ligand average R2eff for the initial value of R2,0. The results of the parameter search are the curves in Fig. 4e and f and Fig. 6f. The numerical values are listed in Table 3. The cis-trans isomerization exchange rate fits the methyl T5-Cγ, V7-Cγ1 profiles and P6-Cδ methylene profile remarkably well. Moreover, we find that the minor state fractional population pB is ~ 11 ± 2%. This percentage is in excellent agreement with the minor cis populations of the Cdc25 ligand estimated from integrations of the 1-D 1H spectrum. Hence, in the simplified system consisting of the truncated Pin1-PPiase construct, the 1H–1H EXSY and side chain 13CH3 and 13CH2 dispersion data report on the same exchange process: cis-trans isomerization of the pT-P motif in Cdc25 by Pin1-PPIase.
Fig. 8.
Example of 1H–1H EXSY of 2 mM Cdc25 in the presence of 50µM Pin1-PPIase at 295 K, 16.4 T. The build-up curves and spectra for methyl protons of pT5
Table 3.
Ligand 13CH3 and 13CH2 exchange parameters with Pin1-PPIase at 295 K and 16.4 T
| Carbon | kex (r/s) | pB | Δppm | R2,0 s−1 |
|---|---|---|---|---|
| pT5-Cγ, methyl | 40 ± 2 | 0.11 ± 0.002 | 0.26 ± 0.02 | 7.1 ± 0.1 |
| V7-Cγ1, methyl | 40 ± 2 | 0.11 ± 0.002 | 0.33 ± 0.03 | 4.8 ± 0.1 |
| P6-Cδ, methylene | 39 ± 2 | 0.10 ± 0.002 | 1.05 ± 0.42 | 7.5 ± 0.8 |
The new information provided by the 13C side-chain dispersion that is not available from the 1H–1H EXSY data are the 13C chemical shift differences between the cis and trans conformers. The 13C chemical shifts are sensitive to local structure; hence, the dispersion data provides potential insight into the side-chain conformational transitions needed for cis-trans isomerization.
Note that we do not directly observe minor cis 13C methyl cross-peaks in the 2-D 13C–1H dispersion spectra—they are “invisible” (Mulder et al. 2001a). This is likely due to the sensitivity associated with natural abundance 13C, and the expected greater broadening of the minor species in a two-state exchange scenario that is slow on the chemical shift time scale (Ishima and Torchia 1999; Millet et al. 2000). The ability of these dispersion measurements to provide information concerning the side-chain 13C chemical shifts—site specific conformational parameters—sheds new light on the exchange process.
In summary, the results from the truncated Pin1-PPIase construct (Fig. 4e, f, and Fig. 6f) strongly suggest that the dispersion profiles for Cdc25 in the presence of full-length Pin1 (Fig. 4a–d, Fig. 6a–d.) reflect more than one exchange process. In essence, the dispersion data from the truncated Pin1-PPIase construct exposes an otherwise “invisible” exchange process masked within the dispersion profiles obtained from full-length Pin1. Thus, the exchange parameters obtained for these profiles are meaningful if the exchange behavior is that of a quasi-two-state process; such a scenario is conceivable if the constituent exchange processes have very different kex values (Grey et al. 2003). This is likely the case here since cis-trans isomerization and binding rates differ by at least an order of magnitude.
Concluding remarks
The results of this work show that 13CH3 and 13CH2 relaxation dispersion studies can give insights into the µs-ms conformational transitions (reorganization) required of a ligand for biological activity. In particular, via studies of the interaction between Pin1 and its Cdc25 ligand, we have shown that the dispersion studies can give insight into the ligand motions necessary for protein complex formation and catalysis The resulting dispersion data yields site-specific exchange rate constants, populations, and critically, 13C chemical shift differences, which are in principle sensitive to changes in the ligand conformational ensemble.
A caveat should be noted: while side-chain 13C chemical shifts are sensitive to local conformation (e.g., side-chain torsion angles), environmental factors (e.g., electric fields, ring current shifts, and changes in solvent exposure) can also play a role. From the dispersion data alone, it is not obvious whether conformational or environmental influences dominate. Recent computational and solid-state NMR studies lend support for the conformational dependence for methyl 13C shift changes (Hong et al. 2009; London et al. 2008). Nevertheless, a conformational interpretation of our side chain 13C relaxation dispersion requires caution. Complementary dispersion experiments of the Pin1 catalytic site, as well as ligand NOESY experiments should help shed light on the dominant chemical shift effects.
From the narrower perspective of understanding the Pin1/Cdc25 ligand interaction, clearly more work remains to be done. For example, we must further measure Cdc25 ligand dispersion in the presence of Pin1-WW domain. Combining such studies with those of the isolated Pin1-PPIase domain above should help enable a more quantitative deconvolution of the dispersion data seen in the presence of full-length Pin1, and help rationalize the apparent differences between backbone and side chain exchange parameters. Such work is in progress. Nevertheless, to the extent that the chemical shifts are dominated by conformational effects, the data presented thus far clearly shows that functionally relevant conformational transitions of the ligand can be described by side-chain relaxation dispersion.
In our study, we have focused on a flexible, peptidic ligand with modest (µM) binding affinity. Such ligands are common in the early stages of drug discovery. What about tight binding (e.g., nano-molar) ligands? In this case, the concentration of the ligand is that of the target protein. Hence, the above dispersion experiments would require either higher protein concentrations ( ~ 500 µM), or isotope-labeled ligands. While we pointed above to the typical paucity of labeled ligands in pharmaceutical research settings, it is also true that if the ligand actually is a tighter binder, then there is likely to be greater interest in taking extra measures to synthesize labeled ligands.
Flexible drug-like molecules typically adopt bound conformations that do not correspond to the conformational energy minima of the free state (Perola and Charifson 2004). This means complex formation introduces new intermolecular interactions that distort conformational energy landscape of the ligand. In turn, this distortion drives conformational reorganization of the ligand upon complex formation. Such re-organization can be thought of as “induced fit” if the free ligand conformational ensemble lacks the bound conformer(s), or “conformational selection” if the free ligand ensemble already fleetingly samples its bound conformer(s). In either scenario, the 13CH3 and 13CH2 relaxation dispersion studies can serve as tools to describe the underlying conformational transitions associated with this re-organization.
We are particularly encouraged by the effectiveness of the 13CH3 dispersion measurements. In typical pharmaceutical research settings, isotope-enriched ligands are a rarity due to cost and syntheses considerations, and this elevates the necessity for 13C experiments to be performed at natural abundance. The high sensitivity afforded by the methyl moiety is well-suited to such natural abundance 13C work. Moreover, methyls are among the most prevalent side chains in drug-like molecules (Bemis and Murcko 1999). Hence, the methyl dispersion experiments should find applicability to a broad range of ligand classes relevant for drug research.
Finally, while our illustration has focused on the Pin1/Cdc25 system,we stress that this strategy is relevant for ligand re-organization problems, in general. In fact, by combining these side-chain 13C dispersion parameters with those of the backbone (Zintsmaster et al. 2008), it should be possible to get a more detailed picture of the conformational distortions the ligand undergoes upon formation of the protein-ligand complex. This may be particularly useful when the three-dimensional structure of the drug-target is unavailable, and therefore, ligand-based approaches focused on pharmacophore development must be adopted (see e.g., van Drie 2003). Integral membrane proteins, such as GPCRs are a well-known example. Alternatively, ligand re-organization may also be important for membrane binding or membrane permeability. Knowing where and how a flexible ligand undergoes conformational changes as a consequence of crossing biological barriers may suggest design strategies for improving ADME properties and oral bioavailability (Navia and Chaturvedi 1996; Veber et al. 2002). In this context, the 13C dispersion experiments described above can shed light on the site-specific conformational changes required for complex formation and drug delivery and thus, help guide strategies of ligand modification aimed at improving activity.
Acknowledgments
We gratefully acknowledge the National Institutes of Health (NIH-RO1GM083081) for support of this work. We thank Mr. John S. Zintsmaster, Ms. Kimberly A. Wilson, Ms. Bipasha Deb, and Mr. Brian J. McArdle for valuable discussions.
References
- Bemis GW, Murcko MA. Properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39(288):7–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Bemis GW, Murcko MA. Properties of known drugs. 2. Side chains. J Med Chem. 1999;42(509):5–5099. doi: 10.1021/jm9903996. [DOI] [PubMed] [Google Scholar]
- Berjanskii M, Wishart DS. NMR: prediction of protein flexibility. Nat Protoc. 2006;1:683–688. doi: 10.1038/nprot.2006.108. [DOI] [PubMed] [Google Scholar]
- Bevington PR. Data reduction and error analysis for the physical sciences. New York City: McGraw Hill; 1969. [Google Scholar]
- Carr HY, Purcell EM. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys Rev. 1954;94:630–638. [Google Scholar]
- Carver JP, Richards RE. A general two-site solution for the chemical exchange produced dependence of T2 upon the Carr-Purcell Pulse separation. J Magn Reson. 1972;6:89–105. [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci U S A. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Crenshaw DG, Yang J, Means AR, Kornbluth S. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J. 1998;17:1315–1327. doi: 10.1093/emboj/17.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DG, Perlman ME, London RE. Direct measurements of the dissociation-rate constant for inhibitor-enzyme complexes via the T1ρ and T2(CPMG) methods. J Magn Reson Ser B. 1994;104(26):6–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- de Dios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- Ernst RR, Bodenhausen G, Wokaun A. Principles of nuclear magnetic resonance in one and two dimensions, Chapter 9, Section 9.3.1. Oxford: Oxford Science Publications; 1987. [Google Scholar]
- Geen H, Freeman R. Band-selective radiofrequency pulses. J Magn Reson. 1991;93:93–141. [Google Scholar]
- Grey MJ, Wang C, Palmer AG., 3rd Disulfide bond isomerization in basic pancreatic trypsin inhibitor: multisite chemical exchange quantified by CPMG relaxation dispersion and chemical shift modeling. J Am Chem Soc. 2003;125:14324–14335. doi: 10.1021/ja0367389. [DOI] [PubMed] [Google Scholar]
- Hong M, Mishanina TV, Cady SD. Accurate measurement of methyl 13C chemical shifts by solid-state NMR for the determination of protein side chain conformation: the influenza a M2 transmembrane peptide as an example. J Am Chem Soc. 2009;131:7806–7816. doi: 10.1021/ja901550q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Estimating the time scale of chemical exchange of proteins from measurements of transverse relaxation rates in solution. J Biomol NMR. 1999;14(36):9–372. doi: 10.1023/a:1008324025406. [DOI] [PubMed] [Google Scholar]
- King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Labeikovsky W, Eisenmesser EZ, Bosco DA, Kern D. Structure and dynamics of pin1 during catalysis by NMR. J Mol Biol. 2007;367:1370–1381. doi: 10.1016/j.jmb.2007.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London RE, Wingad BD, Mueller GA. Dependence of amino acid side chain 13C shifts on dihedral angle: application to conformational analysis. J Am Chem Soc. 2008;130:11097–11105. doi: 10.1021/ja802729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG., 3rd A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- Lundström P, Vallurupalli P, Religa TL, Dahlquist FW, Kay LE. A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J Biomol NMR. 2007;38:79–88. doi: 10.1007/s10858-007-9149-7. [DOI] [PubMed] [Google Scholar]
- Meiboom S, Gill D. Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instr. 1958;29:688–691. [Google Scholar]
- Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG., III The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J Am Chem Soc. 2000;122:2867–2877. [Google Scholar]
- Mulder FAA, de Graaf RA, Kaptein R, Boelens R. An off-resonance rotating frame relaxation experiment for the investigation of macromolecular dynamics using adiabatic rotations. J Magn Reson. 1998;131:351–357. doi: 10.1006/jmre.1998.1380. [DOI] [PubMed] [Google Scholar]
- Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol. 2001a;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001b;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- Namanja AT. Molecular basis for signal transduction in the bimodular cell-cycle enzyme Pin1. University of Notre Dame: Dissertation; 2009. [Google Scholar]
- Namanja AT, Peng T, Zintsmaster JS, Elson AC, Shakour MG, Peng JW. Substrate recognition reduces side-chain flexibility for conserved hydrophobic residues in human Pin1. Structure. 2007;15:313–327. doi: 10.1016/j.str.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Navia MA, Chaturvedi PR. Design principles for orally bioavailable drugs. Drug Discov Today. 1996;1(17):9–189. [Google Scholar]
- Peng JW. New probes of ligand flexibility in drug design: transferred (13)C CSA-dipolar cross-correlated relaxation at natural abundance. J Am Chem Soc. 2003;125:11116–11130. doi: 10.1021/ja030154p. [DOI] [PubMed] [Google Scholar]
- Perni RB, Chandorkar G, Cottrell KM, Gates CA, Lin C, Lin K, Luong YP, Maxwell JP, Murcko MA, Pitlik J, Rao G, Schairer WC, Van Drie J, Wei Y. Inhibitors of hepatitis C virus NS3.4A protease. Effect of P4 capping groups on inhibitory potency and pharmacokinetics. Bioorg Med Chem Lett. 2007;17:3406–3411. doi: 10.1016/j.bmcl.2007.03.090. [DOI] [PubMed] [Google Scholar]
- Perola E, Charifson PS. Conformational analysis of drug-like molecules bound to proteins: an extensive study of ligand reorganization upon binding. J Med Chem. 2004;47:2499–2510. doi: 10.1021/jm030563w. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spera S, Bax A. Empirical correlation between protein backbone conformation and Ca and Cb 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- van Drie JH. Pharmacophore discovery–lessons learned. Curr Pharm Des. 2003;9:1649–1664. doi: 10.2174/1381612033454568. [DOI] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- Vila JA, Scheraga HA. Factors affecting the use of 13C(alpha) chemical shifts to determine, refine, and validate protein structures. Proteins. 2008;71:641–654. doi: 10.1002/prot.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintjens R, Wieruszeski JM, Drobecq H, Rousselot-Pailley P, Buee L, Lippens G, Landrieu I. 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides. J Biol Chem. 2001;276:25150–25156. doi: 10.1074/jbc.M010327200. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Xu XP, Case DA. Probing multiple effects on 15 N, 13C alpha, 13C beta, and 13C’ chemical shifts in peptides using density functional theory. Biopolymers. 2002;65:408–423. doi: 10.1002/bip.10276. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- Zintsmaster JS, Wilson BD, Peng JW. Dynamics of ligand binding from 13C NMR relaxation dispersion at natural abundance. J Am Chem Soc. 2008;130:14060–14061. doi: 10.1021/ja805839y. [DOI] [PMC free article] [PubMed] [Google Scholar]