Abstract
Prions are fascinating but often misunderstood protein aggregation phenomena. The traditional association of the mammalian prion protein with disease has overshadowed a potentially more interesting attribute of prions - their ability to create protein-based molecular memories. In fungi, prions alter the relationship between genotype and phenotype in a heritable way that diversifies clonal populations. Recent findings in yeast indicate that prions may be much more common than previously realized. Moreover, prion-driven phenotypic diversity increases under stress, and can be amplified by the dynamic maturation of prion-initiating states. We argue that these qualities allow prions to act as bet-hedging devices that facilitate yeast’s adaptation to stressful environments, and may speed the evolution of new traits.
Introduction
Prions are self-replicating protein entities that underlie the spread of a mammalian neurodegenerative disease, variously known as Kuru, scrapie, and bovine spongiform encephalopathy, in humans, sheep and cows, respectively [1]. However, most prions have been discovered in lower organisms and in particular, the yeast Saccharomyces cerevisiae. Despite assertions that these prions, too, are diseases [2] (Box 1), many lines of evidence suggest that these mysterious elements are generally benign and, in fact, in some cases beneficial. In fungi, prions act as epigenetic elements that increase phenotypic diversity in a heritable way and can also increase survival in diverse environmental conditions [3–6]. In higher organisms, prions may even be a mechanism to maintain long-term physiological states, as suggested for the Aplysia californica (sea slug) neuronal isoform of CPEB, cytoplasmic polyadenylation element binding protein. The prion form of this protein appears to be responsible for creating stable synapses in the brain [7]. CPEB is the prominent first example of what may be a large group of prion-like physiological switches, the potential scope of which cannot be given adequate coverage here. Instead, this piece will focus on prions as protein-based genetic elements – their ability to drive reversible switching in diverse phenotypes, and the way that such switching can promote the evolution of phenotypic novelty.
Box 1. The alternative view: prions as diseases.
The yeast prion field is embroiled in controversy over whether or not these protein-based elements of inheritance are simply protein-misfolding “diseases” of yeast, or instead serve important biological functions. This article advocates the latter. Arguments for the former are summarized here.
The yeast prions [PSI+] and [URE3] have not been observed in natural populations. In a screen for the pre-existence of [PSI+], [URE3], and [RNQ+] in a panel of 70 diverse yeast strains, only [RNQ+] was observed [68]. The authors concluded that there is likely to be strong selective pressure against these prions. Indeed, the majority (~75 %) of phenotypes found to be revealed by [PSI+] are detrimental [13]. Nevertheless, these observations are consistent with the proposed functionality of prions as either bet hedging devices or evolutionary capacitors, both of which predict prion states to occur infrequently and to be disadvantageous on average. Since natural selection acts on the geometric mean fitness of a genotype over time, disadvantages suffered by a small fraction of prion-containing cells are easily outweighed by occasional strong advantages [15, 69].
The strain phenomenon of mammalian and yeast prions may result from a lack of positive selective pressure acting on the prion states [2]. Corroborating this idea is the apparent absence of variation in the [Het-s] prion of Podospora anserina, a prion largely accepted to function in the process of heterokaryon incompatibility [2, 70], which limits the mixing of cytoplasm and thus the transfer of deleterious infectious elements, between mycelia. However, the existence of diverse prion strains adds an additional layer of prion-mediated phenotypic heterogeneity, which itself may be under positive selective pressure in the bet hedging and evolutionary capacitance models.
Prion determining regions may have alternative, (non-prion) functions. The prion domains of Sup35 and Ure2 were long thought to be dispensable for the cellular functions of their respective proteins, indicating they may have evolved specifically for prion formation. Some exceptions to this generalization have been discovered [2], and it is possible that these protein regions have alternative, albeit subtle, activities not directly related to prion formation. Prions could then be artifacts of selective pressure for other, non-prion-related activities [2]. However, functional pleiotropy is not uncommon, especially among intrinsically disordered protein regions [71], and is not in itself evidence against positive selective pressure for prion formation.
A related observation is that some prion homologs from other species appear incapable of prion formation [2, 72], suggesting their conservation may not be due to prion formation. An alternative explanation is that prion formation may have been precluded by interspecific differences in trans-acting prion promoting factors like Hsp104 and Rnq1, or differences in natural growth conditions or niche-specific stresses. Nevertheless, the majority of conserved prion domains that have been tested do retain the ability to form prions [37–39, 72], despite the existence of numerous possibilities for mutational inhibition of prion formation. Finally, it is possible or even probable that prion formation evolves within the context of Q/N-rich sequences having alternative cellular functions as protein-protein interaction domains. Restricted conservation of prion formation may indicate that prion formation in these cases is a relatively recent exaptation.
The self-templating replicative state of most biochemically characterized prions is amyloid [5, 8] (Figure 1), although other types of self-propagating protein conformations may also give rise to prion phenomena [9, 10]. Amyloid is a highly ordered, fibrillar protein aggregate with a unique set of biophysical characteristics that facilitate prion propagation: extreme stability, assembly by nucleated polymerization, and a high degree of templating specificity. Prion propagation proceeds from a single nucleating event that occurs within an otherwise stable intracellular population of non-prion conformers. The nucleus is then elongated into a fibrillar species by templating the conformational conversion of non-prion conformers [11, 12] (Figure 1). Finally, the growing protein fiber fragments into smaller propagating entities, which are ultimately disseminated to daughter cells [6]. Because the change in protein conformation causes a change in function, these self-perpetuating conformational changes create heritable phenotypes unique to the determinant protein and its genetic background (Figure 1). The genetic properties that arise are distinct from those of most nuclear-encoded mutations: prion phenotypes are dominant in genetic crosses and exhibit non-Mendelian inheritance patterns. Hence prion-based genetic elements are denoted with capital letters and brackets – “[PRION]”.
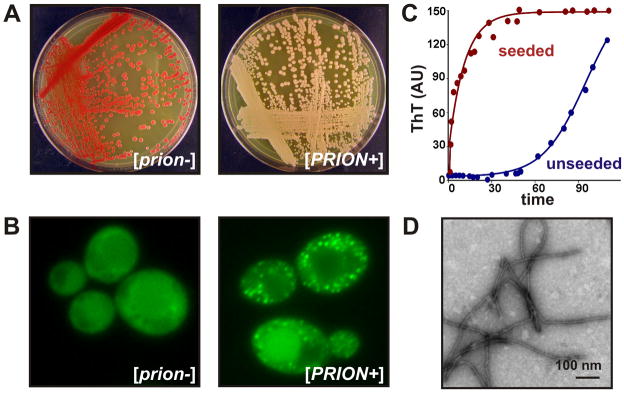
Figure 1. Prions as self-templating aggregates.
(a) Prions of S. cerevisiae cause heritable changes in phenotype. In this particular genetic background, the prion [PSI+] can be observed by white coloration and adenine prototrophy due to translational readthrough of a nonsense mutation in the ADE1 gene. However, the cryptic genetic variation that can be revealed by [PSI+] is inherently polymorphic resulting in a wide variety of strain-specific [PSI+] phenotypes [3].
(b) Prion phenotypes are generally caused by a reduction of the prion protein’s normal cellular activity. In vivo, the aggregation and partial loss-of-function of the prion protein, can be observed by the presence of Sup35-GFP foci in [PSI+] cells. These foci are composed of self-templating prion aggregates that are cytoplasmically transmitted during cell division.
(c) Nucleated aggregation of a prion protein. Purified prion protein populates a soluble state for an extended period of time, then polymerizes exponentially after the appearance of amyloid nuclei (blue trace). The lag phase can be eliminated by the addition of small quantities of preformed aggregates (red trace), demonstrating the biochemical property underlying the self-propagating prion state [11].
(d) The self-propagating prion conformation is amyloid-like, as seen by the highly ordered, fibrillar appearance of prion domain aggregates visualized by transmission electron microscopy. Amyloid is a one-dimensional protein polymer. Its free ends template a protein folding reaction that incorporate new subunits while regenerating the active template with each addition.
Protein remodeling factors, chaperones, and other protein quality control mechanisms interact with prions at every step in their propagation. Further, prion-driven phenotypic switches are modulated by environmental conditions that perturb protein homeostasis [13] – the proteome-wide balance of protein synthesis, folding, trafficking, and degradation processes [14]. Prions could thereby constitute an intrinsic part of the biological response to stress. We postulate that the relationship between prions and protein homeostasis, as well as the dynamic nature of prion propagation, render prions into sophisticated evolutionary bet-hedging devices. Herein, we explore multiple intriguing features of prion biology that together argue for a general role for prions in adaptation to new environments, and thereby the evolution of new traits.
Prions as bet-hedging devices
Prions can allow simple organisms to switch spontaneously between distinct phenotypic states [4]. For this reason, prions can be regarded as bet-hedging devices. Bet-hedging devices increase the reproductive fitness of organisms living in fluctuating environments by creating variant subpopulations with distinct phenotypic states [15] (Box 2).
Box 2. Phenotypic heterogeneity and bet-hedging strategies in microorganisms.
Individual cells in isogenic microbial cultures often exhibit a high degree of phenotypic heterogeneity under homogenous culture conditions [44]. Some of the best characterized examples of heterogeneously expressed phenotypes include cellular differentiation in Bacillus subtilis, lactose utilization, chemotaxis and antibiotic persistence in E. coli, surface antigen expression in the malaria parasite Plasmodium falciparum, and the expression of cell wall adhesins that control flocculation and invasive growth in baker’s yeast [44]. Stochasticity or noise in gene expression is generally believed to be an important contributor to cell individuality but other drivers of phenotypic heterogeneity have been proposed, including the cell cycle, ageing, biological rhythms, individual cell growth rates and metastably inherited epigenetic changes [44].
Such population-level heterogeneity can increase an organism’s ability to survive in fluctuating environments. Fluctuations can involve the macro-environment, the external micro-environment, and even the internal cellular environment. Phenotypic heterogeneity is an unavoidable result of biological noise, but can also result from adaptive phenotypic variation, or “bet-hedging” [15, 73]. Classic examples of bet-hedging include growth decisions like insect diapause and seed dormancy [15], but stochastic switching between phenotypes in microbes can similarly increase mean fitness over multiple generations and thus benefits the bet-hedging genetic lineage [44, 73]. Bet-hedging can be a theoretically viable alternative to phenotypic switching based on environmental sensing [74], and can be experimentally evolved in bacteria [73]. The value of stochastic switching is shaped by many factors, including the frequency, predictability, and severity of environmental change; the capacity of the organism to respond directly to change; and the inherent evolvability of the population [15, 74].
The first prion protein proposed to increase survival in fluctuating environments is the translation termination factor Sup35, which forms a prion state called [PSI+] [4]. This prion reduces Sup35 activity relative to the non-prion, or [psi−] state, thereby creating a variety of phenotypes related to alterations in translation fidelity [3, 16–18]. A surprisingly large fraction of the phenotypes (~25% in one study [13]) are advantageous under particular growth conditions. While reduced translational fidelity can not, in the long run, be advantageous for growth, in the short run changes in gene expression brought about by [PSI+] can allow cells to grow in the presence of antibiotics, metals and other toxic conditions, or with different carbon or nitrogen sources, depending on the genetic background. Because cells spontaneously gain the prion at an appreciable frequency (10−7 to 10−6) [19–21], at any one time a sizable population of yeast cells will contain a few that have already switched states. If the environment is such that [PSI+] is beneficial, these cells would then have a greater chance to survive in that environment. Importantly, the prion state can be reversed by its occasional loss during cell division [22] (with as yet undetermined frequencies), resulting in progenitors with the original [psi−] phenotype. If after a period of growth, the environment changes to a state where [PSI+] is not advantageous, those few cells that have spontaneously lost the prion then have a survival advantage. From a gene-centric point of view, the net effect of this phenotype switching is that the common genotype shared by both [PSI+] and [psi−] cells survives through the strenuous series of environmental transitions. Even if the rare switches to the [PSI+] state are commonly disadvantageous, [PSI+] could dramatically improve the long-term fitness of a genotype if it is advantageous on occasion. Related phenotypic switching phenomena, like the reversible appearance of antibiotic-resistant “persister” bacteria, appear to constitute environmentally-optimized risk-reduction strategies [23] (Box 2).
Other than Sup35, the best characterized yeast prion is the Ure2 nitrogen catabolite repressor. Its prion state, [URE3], causes cells to constitutively utilize poor nitrogen sources [6]. This same phenotype, when conferred by URE2 loss-of-function mutants, has been shown to confer a proliferative advantage to cells in fermenting grape must [6], strongly suggesting that this prion, too, may have a functional role in coping with yeast’s diverse ecological niches.
Until recently, the prion field has been confined to a small handful of proteins, and for this reason, conjectures about their potential roles in adaptation and evolution have been limited. However, a wave of recent discoveries in yeast has dramatically expanded the prion world as we know it (Table 1). The newly discovered prions include functionally diverse proteins: multiple chromatin remodeling and transcription factors [5, 24, 25], a metacaspase [26], and a range of additional prionogenic proteins whose putative endogenous prion states are yet to be examined [5]. We suggest that the existence of these prions and the phenotypic heterogeneity they produce contributes to a general bet-hedging strategy that arms yeast populations against environmental fluctuations. Recent analyses of some of these novel prions lend support to this idea [5, 25].
Table 1.
Known and candidate prions
| Prion determinant | Prion state | Organismb | Protein function | Consequences of prion state | Reference |
|---|---|---|---|---|---|
| Nativea prions | |||||
| PrP | PrPSc | Mammals | Neuronal growth and maintenance, hematopoietic stem cell renewal | Neurodegeneration and death | [1] |
| Ure2 | [URE3] | S. cerevisiae and related yeasts | Represses transcription of nitrogen catabolic genes | Indiscriminate utilization of nitrogen sources | [78] |
| Sup35 | [PSI+] | S. cerevisiae and related yeasts | Translation termination | Increased nonsense suppression, translational frameshifting, changes in mRNA stability | [78] |
| Rnq1 | [PIN+] | S. cerevisiae | Unknown | Increased appearance of other prionsc | [79] |
| HET-s | [Het-s] | P. anserina | Heterokaryon incompatibility | Inhibits fusion between [Het-s] and het-S mycelia | [70] |
| Swi1 | [SWI+] | S. cerevisiae | Transcription regulation | Altered carbon source utilization | [24] |
| Mca1 | [MCA] | S. cerevisiae | Regulation of apoptosis, cell cycle progression | Unknown | [26] |
| Cyc8 | [OCT+] | S. cerevisiae | Transcription repression | Altered carbon source utilization, flocculation | [25] |
| Mot3 | [MOT3+] | S. cerevisiae | Transcription regulation | Altered cell wall composition | [5] |
| Pma1/Std1 | [GAR+] | S. cerevisiae | plasma membrane proton pump (Pma1) and glucose signaling (Std1) | Indiscriminate utilization of carbon sources | [10] |
| Candidate prions | |||||
| CPEB, neuronal isoformsd | - | A. californicac | Translation regulation of synapse-specific mRNAs | Localized protein synthesis at activated synapses; maintains long term facilitation | [7] |
| ? | [ISP+] | S. cerevisiae | - | Antisuppression of nonsense suppressors | [80] |
| 19 other proteinse | - | S. cerevisiae | Diverse | Undetermined | [5] |
Prions that have been shown to propagate in the native host and whose causal protein has been identified
Limited to organisms that have been examined for the particular prion.
To varying extents, likely to be a general property of prions.
Prion properties were examined in a non-native host, S. cerevisiae.
Proteins contain regions that can propagate as prions when fused to a reporter domain.
S. cerevisiae, Saccharomyces cerevisiae; P. anserina, Podospora anserina; A. californica, Aplysia californica.
[MOT3+] is a prion formed by the transcription factor Mot3, an environmentally responsive regulator of yeast cell wall composition and pheromone signaling [27, 28]. In general, the cell surface of yeast determines the communication and interaction of yeast cells with the environment, yet it is also involved in a host of morphological and behavioral phenotypes, such as cell growth, cell division, mating, filamentation, and flocculation. Whether the phenotypic variation introduced by [MOT3+] affects all of these processes remains to be explored, but [MOT3+] does confer increased resistance to certain cell wall stressors [5]. Therefore, the phenotypes produced by [MOT3+] should be advantageous in many microbial environments. The biological significance of Mot3 prion formation is supported by its high frequency of appearance – approximately 1 in 10,000 cells ([5] and Halfmann and Lindquist, unpublished observation).
[SWI+] and [OCT+] are formed by the globally acting transcriptional regulators, Swi1 and Cyc8, respectively [24, 25]. [SWI+] cells are resistant to the microtubule disruptor, benomyl [5]; and [OCT+] induces flocculation [25], a growth form that has been shown to protect cells from diverse stresses [29]. Given the large size and complexity of the gene networks regulated by each of these prion transcription factors, it is likely that many more phenotypes are yet to be linked with prions.
Finally, for the well-characterized prions, it has been established that the presence of one protein in its prion state can influence the prion switching of other proteins. The [RNQ+] prion, for instance, strongly increases the rate of appearance of other prions [5, 6]. Conversely, some prions destabilize each other when both exist in the same cell [30]. Such prion cross-talk is influenced both by the sequence similarity between the proteins and the degree to which they share common components of the cellular prion-propagating machinery [31, 32]. The likely existence of over twenty interconnected prion switches [5], all contributing to phenotypic heterogeneity, would greatly increase a genetic lineage’s potential to explore phenotypic space. Prions are being discovered at an increasingly rapid pace, suggesting that many exciting possibilities remain to be discovered en route to a deeper understanding of the prevalence and functionality of prions in biology.
Prions as evolutionary capacitors
In addition to “normal” bet-hedging, prions may have an even deeper and more sophisticated role in microbial evolution. Specifically, prions have been proposed to be capable of evolutionary capacitance [6]. An evolutionary capacitor is any entity that normally hides the effects of genetic polymorphisms, allowing for their storage in a silent form, and releases them in a sudden stepwise fashion [33]. The complex phenotypes produced by the sudden expression of accumulated genetic variation on occasion will prove beneficial to the organism. As the organism proliferates, further genetic and epigenetic variations will accumulate that stabilize the beneficial phenotype. The extent to which evolutionary capacitors impact the evolution of natural populations is highly debated, and even more so the notion that capacitance itself can be subject to natural selection [34].
However, the accumulated evidence that at least one prion protein, Sup35, acts in this manner is exceedingly difficult to dismiss. Sup35 can act as an evolutionary capacitor by connecting protein folding to the relationship between genotype and phenotype in a remarkable way. The reduced translation fidelity brought about by Sup35’s prion state, [PSI+], results in the translation of previously silent genetic information through a variety of mechanisms including stop-codon readthrough and ribosome frameshifting [3, 16, 35, 36]. Stop-codon readthrough can also affect genetic expression by changing mRNA stabilities. Untranslated regions and cryptic RNA transcripts experience relaxed selection under normal ([psi−]) conditions, and consequently, are free to accumulate genetic variation. Upon the appearance of [PSI+], these polymorphisms become phenotypically expressed. Because [PSI+] operates on genetic variation in a genome-wide fashion, it allows for the sudden acquisition of heritable traits that are genetically complex [3]. Such traits are initially unlikely to become [PSI+]-independent because they involve multiple genetic loci and cells will revert to their normal phenotype when they lose the prion. But if the environment that favors the changes in gene expression brought about by [PSI+] occurs frequently or lasts for a very long time, as the population expands, mutations will accumulate that allow cells to maintain the traits even when they revert to normal translational fidelity through the spontaneous loss of [PSI+]. Arguing that Sup35 is under selective pressure to maintain the ability to reveal such variation, Sup35 homologs from other yeasts have conserved prion-forming capabilities, despite their sequences having diverged extensively over hundreds of millions of years [37–39]. Mathematical modeling confirms that the complexity of [PSI+]-revealed phenotypes can theoretically account for the evolution of its prion properties in yeast [40]. Finally, a phylogenetic analysis of the incorporation of 3′ untranslated regions (UTRs) into coding sequences provides compelling evidence for [PSI+]-mediated evolution in natural yeast populations. When comparing yeast and mammalian genomes, yeast displayed a strong bias for mutation events leading to in-frame, rather than out-of-frame incorporation of 3′ UTRs [41]. Thus, yeast 3′ UTRs are translated at a relatively high frequency, consistent with the occasional appearance of [PSI+] in natural populations.
Buffering of phenotypic variation is an inherent property of regulatory networks, such that the conditional reduction of network integrity may be a common mode of evolutionary capacitance [33]. The distinction between this type of capacitance and prions is that the latter are necessarily epigenetic, and therefore provide a mechanism for the persistence, and ultimately, genetic assimilation, of the revealed phenotypes [33]. Prion-associated phenotypes can appear spontaneously and persist for multiple generations, whereas the revelation of variant phenotypes by other capacitors is generally contingent on stress, and consequently, relatively transient.
Is prion-driven evolutionary capacitance unique to Sup35, or might prion formation within any number of proteins also promote the expression of hidden genetic variation? Intriguingly, many of the newly identified prions are situated to function as genetic capacitors in their own right. Conspicuously overrepresented among these prionogenic proteins are gene products that control gene expression, cell signaling and the response to stimuli such as stress ([5] and Table 1). Many of them represent highly connected nodes in the yeast genetic network. The Swi1 chromatin remodeler, for instance, regulates the expression of 6% of the yeast genome [24]. Likewise, Cyc8 represses 7% of the yeast gene complement [42]. The prion candidates Pub1, Ptr69 and Puf2 are members of a family of RNA-binding proteins that regulate the stability of hundreds of mRNAs encoding functionally related proteins [43]. The strong enrichment of putative prions among proteins that regulate and transact genetic information suggests that prion-based switches evolve preferentially among proteins whose functions impinge on multiple downstream biological processes. Pre-existing genetic polymorphisms whose expression is altered by these prions would create different phenotypes in different genetic backgrounds. Thus, many prions are quite likely to create strong and complex phenotypes upon which natural selection can act.
Prion formation as an environmentally responsive adaptation
Many bet-hedging devices are environmentally responsive [44] (Box 2). That is, in addition to entirely stochastic switches, organisms may also make what, in effect, amounts to “educated guesses” by integrating environmental cues to modulate the frequency of phenotypic switching. Indeed, the frequency of prion switching is affected by environmental factors. The appearance of [PSI+] is strongly increased by diverse environmental stresses [13, 45]. Incidentally, this property is necessary and sufficient for [PSI+] formation to have been favored by natural selection for evolvability [21]. Other well-characterized prions are also known to be induced by prolonged refrigeration and/or deep stationary phase [46]. Because prions are a special type of protein misfolding process, logically their induction is intrinsically tied to environmental stresses that perturb protein stability. Many if not most polypeptides have a generic capacity to form amyloid [47]. Situations that alter native protein stability, like thermal stress, altered pH, or metal ion imbalances, are therefore likely to facilitate polypeptides’ access to prion or prion-like amyloid conformations [47] with the potential to perpetuate phenotypic changes even after the stress subsides.
The connection to environmental stresses is much deeper than that, however. Protein quality control machinery is ubiquitous throughout all kingdoms of life and is essential for both normal protein folding and for coping with stress. Components of the ubiquitin-proteasome system strongly impact prion formation [46]. And prion propagation requires the actions of members of the Hsp40, Hsp70, and Hsp110 chaperone families as well as the AAA+ protein disaggregase Hsp104 [46, 48]. Hsp104 is a member of ClpA/ClpB family of chaperones whose members are found throughout bacteria, fungi, plants and eukaryotic mitochondria. Hsp104 provides thermotolerance by resolubilizing stress-induced protein aggregates, and also has the unique ability to sever amyloid fibers into new prion propagons. This property has been conserved for hundreds of millions of years of fungal evolution [49]. On the other hand, the Hsp104 protein of fission yeast appears incapable of propagating amyloid-based prions, despite maintaining its important ability to solubilize non-amyloid stress-induced protein aggregates [50]. We note that fission yeast also has a relative paucity of computationally predicted prions [51], consistent with the suggestion that Hsp104’s amyloid shearing capability coevolved with prions to promote their propagation. Indeed, at least 25 of the 26 known amyloidogenic yeast prion domains require Hsp104 for their propagation as prions [5, 26, 52].
Perhaps the dominant force, then, for stress-induced prion formation involves perturbations in the interactions of prion proteins with chaperones and the cellular environment. The distribution of proteins between soluble and aggregated states is exquisitely sensitive to the status of the protein homeostasis network, which comprises protein synthesis, folding, sorting, and degradation machinery [53]. Chaperones are highly connected in protein interaction networks and serve an important role as transducers of the stress response [53]. Prion proteins, in turn, are highly connected to chaperones and thus to the protein homeostasis network at large. Prion conformational switching may therefore respond to stress indirectly through, for example, alterations in the abundance, availability, and connectivity of chaperones like Hsp104 and Hsp70s [54]. The induction of prions by diverse proteostatic stresses, and their dependence on chaperones for propagation, may reflect the long history of chaperone involvement in the relationship between environment and phenotype.
Phenotypic diversity further enhanced by prion conformational and temporal diversity
The morphological adaptive radiation of organisms appears to result predominantly from genetic changes that have quantitative rather than qualitative effects [55]. In yeast and other microbes, social behaviors like mating, flocculation, and colony formation are subject to frequent stochastic changes in the expression of extracellular adhesins, leading to the rapid divergence of variant subpopulations [56]. These changes facilitate their expansion into diverse and highly dynamic ecological niches. The mechanisms for such changes are both genetic and epigenetic in nature [33, 56], and include nucleotide repeat expansions and contractions, chromatin remodeling, and as recently discovered, prion formation [25]. Importantly, all of these mechanisms tend to modulate the activity levels, rather than the functional nature of, the affected gene products.
The ability of organisms to explore such modulations of gene activity, either as individuals (e.g. phenotypic plasticity), or as members of a genetic lineage (e.g. bet-hedging), enhances their survival under adverse conditions and is thought to facilitate the subsequent genetic assimilation of beneficial phenotypic variations [33]. Molecular mechanisms that allow for the rapid stabilization or amplification of initially non-genetic adaptive phenotypes within a lineage could greatly accelerate this process. Indeed, epigenetic processes are likely to play an important role in adaptive diversification [57]. As examined below, prions may represent an ideal epigenetic mechanism for the heritable modulation of gene activity.
Prions have a unique capacity to stratify protein functionality into multiple semi-stable levels, which greatly increases the phenotypic diversity created by prion-driven switches. It derives from the unusual and variable way in which prion conformers nucleate and propagate, and has both static and temporal components. For a given prion, multiple distinct yet related protein conformations can each self-perpetuate (Figure 2a). These prion “strains” differ in phenotypic strength and heritability. Strain multiplicity has been observed with both mammalian and yeast prions [12], and is a common feature of diverse amyloids when polymerized in vitro [58]. The nature of the conformational differences between strains is still poorly understood, although progress has been made in elucidating how physical differences between amyloid strains – such as the extent of sequence involved with the fibril core of the amyloid – translate into differences in amyloid growth and division rates, and in turn the phenotypic strength of the prion [12]. Importantly for the bet-hedging aspect of prion biology, the conformational plasticity of the prion nucleation process further increases the phenotypic “coding potential” of a single prion gene.
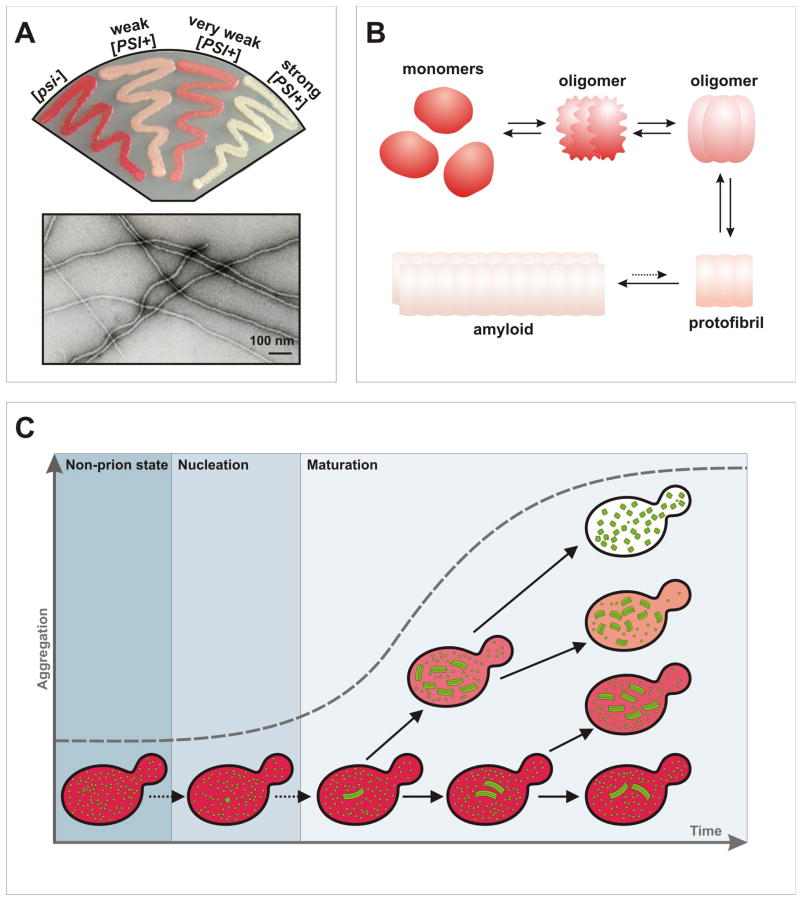
Figure 2. Conformational and temporal diversity of prion states.
(a) Prions create multiple stable phenotypic states, or “strains”. [PSI+] strains differ by their levels of nonsense suppression, with stronger strains having less functional Sup35 available to fulfill its role in translation termination, giving rise to a whiter coloration in a particular genetic background (top). At the molecular level, strains are determined by amyloid conformational variants (bottom) that arise during nucleation but then stably propagate themselves.
(b) Along with the conformational diversity apparent in the end products of amyloid formation, multiple conformational variants are also transiently populated during the early stages of amyloid assembly, and may constitute integral on-pathway species [47]. These oligomeric intermediates likely have limited self-templating capacity, but nevertheless may contribute to the weak phenotypes associated with incipient prion states.
(c) Incipient prion states acquire progressively stronger phenotypes and stabilities, possibly via mass-action population dynamics of prion particles. A number of elegant studies have correlated the phenotypic strength of the prion state with the intracellular number of prion particles [75, 76]. Upon de novo nucleation within a prion-free cell, prion polymerization onto limiting fiber ends proceeds during the “maturation” phase under pre-steady state conditions. Upon each cell division, prion particles are distributed passively and asymmetrically to daughter cells [22]. Progeny that inherit more particles will have faster total prion polymerization rates and correspondingly stronger phenotypes, and will tend to accumulate more prion particles that will in turn strengthen the prion phenotype in subsequent generations (light pink and white cells). Conversely, cells that inherit fewer particles will have slower polymerization rates and weaker phenotypes (red and pink prion-containing cells), and themselves will tend to accumulate fewer particles to pass on to their progeny. Such noise in prion distribution may allow prions to stratify protein functionality along a continuum of semi-stable phenotypes (e.g. red cells, pink cells, and white cells) within a small number of cell generations.
Several observations also demonstrate a temporal component to the strength and stability of prion phenotypes. For example, the mitotic stability of newly induced prion states increases with repeated cell passaging [37, 59–62]. Additionally, selection for incipient prions using mild selective conditions creates a much larger population of strong prion states than would be expected from the numbers obtained by immediate stringent selection [13, 63–65]. Recent observations that even “non-prion” amyloids, such as polyglutamine-based aggregates, can become mitotically stable [66], suggest that a capacity for the maturation of propagating states may be a generic feature of amyloid-like aggregates. The rate of prion maturation is strongly influenced by Hsp104 activity [66], indicating an additional mode by which the protein homeostasis network connects the environment to epigenetic changes.
Multiple mechanisms for generating prion diversity temporally can be envisioned (Figure 2), including amyloid strain-like conformational transitions [66], the mass-action population dynamics of prion particles, the variable association of prion particles with specific cellular structures, and the participation in early stages of prion propagation by an array of oligomeric species that have been increasingly observed en route to amyloid fibrillation [11, 12, 67]. It is plausible that some pre-amyloid species have rudimentary self-propagating activities themselves.
Regardless of the mechanisms involved, what is clear is that incipient prion states represent dynamic molecular populations, a view that challenges the prevailing assumption that prions increase phenotypic heterogeneity solely by acting as simple binary switches. Prion nucleation allows for a single protein species to create a dynamic continuum of semi-stable phenotypes (Figure 2c) that do not require genetic, expression-level, or posttranslational regulatory changes to that protein. For each prion protein, natural selection could operate at any point in this continuum to favor prion-containing cells, resulting in their clonal expansion relative to other cells and acting to shift the distribution of phenotypes within the continuum. Stress-induced formation of prions followed by their iterative maturation offers a rapid route to tunable, advantageous phenotypes. Ultimately, the beneficial phenotypes conferred by prions can become hard-wired by the accumulation of genetic and further epigenetic modifications [4]. In this way, semi-stable phenotypic heterogeneity conferred by the diversity of prion conformations and maturation states would greatly improve the odds of organismal survival in unpredictable or fluctuating environments, and thereby facilitate subsequent adaptive genetic changes.
Concluding remarks
The ability of prions to create heritable phenotypic diversity that is inducible by stress, coupled with the conformational and temporal diversity of prion states, suggests a prominent role for prions in allowing microorganisms to survive in fluctuating environments. However, broader validation is needed, and many questions remain (Box 3). The field of prion biology is now poised to answer these questions, and in so doing, make important contributions to our understanding of evolutionary processes. In particular, we may more fully realize that organisms have specific mechanisms to enhance the evolution of phenotypic novelty.
Box 3. Outstanding questions and future directions.
Many important questions remain to be answered before a comprehensive understanding can be achieved about prions’ roles in biology.
How frequently do prions occur in natural populations?
Random sampling among natural yeast populations has revealed few stably perpetuating prion states [68]. However, a more informative experiment might be to examine natural strains for the transient appearance of prion states. For example, one might introduce fluorescent protein reporters of prion states [77] into natural yeast strains and follow population dynamics of prion-containing vs. prion-free cells under diverse growth conditions.
Are prion states generally inducible by stress?
Early steps have succeeded in establishing a definitive link between environmental stress and prion formation [13]. But whether stress-induced prion formation results from positive selective pressure on prions, or instead reflects the general decrease in protein quality control under stress remains to be determined. With improved prion reporters and an expanding set of diverse prions to study (Table 1), it is now feasible to elucidate whether there is specificity among prions in response to diverse stresses, and whether such specificity can be attributed to selective pressures acting on prion-encoded phenotypes.
Can the dynamics of prion states contribute to, and explain, their biology?
The technical limitations of genetic and cell biological approaches have precluded detailed characterizations of the early, sub-phenotypic stages of prion propagation in vivo. The molecular events giving rise to prion nucleation and the ultimate emergence of a stable prion state are virtually unknown. Mathematical modeling approaches, coupled with advances in microscopic imaging, have the potential to contribute enormously toward filling this gap. This, in turn, could improve estimates of true prion switching rates and perhaps expedite the discovery of novel prion-like processes. Additionally, the accurate quantitation of the rates of prion appearance and disappearance will be critical for the modeling and validation of proposed bet hedging-related functions of prions.
Do prions occur, and with what consequence, in other organisms?
For historical and practical reasons, our knowledge of the world of prions is shaped heavily by experimentation in yeast. But if prions are functional in the sense laid out in this Opinion, we might expect them to occur commonly in other organisms. Particularly, might there be underlying prion determinants for some of the phenotypic switches commonly observed in bacteria [44]? On the other hand, if prions are found to be largely fungal or even S. cerevisiae phenomena, what might we learn from the diversity of evolutionary strategies to deal with protein aggregation?
Acknowledgments
We are grateful to Sebastian Treusch for fruitful discussions, and members of the Lindquist lab for critical reading of the manuscript. RH was supported by a fellowship of the National Science Foundation (NSF). SA was supported by a research fellowship of the Deutsche Forschungs-gemeinschaft (DFG), and the G. Harold & Leila Y. Mathers Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aguzzi A, et al. The prion’s elusive reason for being. Annu Rev Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 2.Wickner RB, et al. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.True HL, et al. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 4.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 5.Alberti S, et al. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 7.Si K, et al. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 8.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 9.Wickner RB, et al. Yeast prions: evolution of the prion concept. Prion. 2007;1:94–100. doi: 10.4161/pri.1.2.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23:2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 12.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyedmers J, et al. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 15.Seger J, Brockmann HJ. What is bet-hedging? Oxford Surv Evol Biol. 1987;4:192–211. [Google Scholar]
- 16.Liebman SW, Sherman F. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. J Bacteriol. 1979;139:1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namy O, et al. Epigenetic control of polyamines by the prion [PSI+] Nat Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- 18.Patino MM, et al. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 19.Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 20.Tank EM, et al. Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability. Mol Cell Biol. 2007;27:5445–5455. doi: 10.1128/MCB.02127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancaster AK, et al. The Spontaneous Appearance Rate of the Yeast Prion [PSI+] and Its Implications For the Evolution of the Evolvability Properties of the [PSI+] System. Genetics. 2009 doi: 10.1534/genetics.1109.110213. ( http://www.genetics.org) [DOI] [PMC free article] [PubMed]
- 22.Cox BS, et al. Reversion from suppression to nonsuppression in SUQ5 [psi+] strains of yeast: the classificaion of mutations. Genetics. 1980;95:589–609. doi: 10.1093/genetics/95.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kussell E, et al. Bacterial persistence: a model of survival in changing environments. Genetics. 2005;169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Z, et al. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel BK, et al. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemecek J, et al. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci U S A. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Grishin AV, et al. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics. 1998;149:879–892. doi: 10.1093/genetics/149.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abramova N, et al. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J Bacteriol. 2001;183:2881–2887. doi: 10.1128/JB.183.9.2881-2887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smukalla S, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradley ME, et al. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathur V, et al. Ssa1 overexpression and [PIN(+)] variants cure [PSI(+)] by dilution of aggregates. J Mol Biol. 2009;390:155–167. doi: 10.1016/j.jmb.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pigliucci M. Is evolvability evolvable? Nat Rev Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- 35.Namy O, et al. Epigenetic control of polyamines by the prion [PSI(+)] Nat Cell Biol. 2008;10:1069–75. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- 36.Wilson MA, et al. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc Natl Acad Sci U S A. 2005;102:10244–10249. doi: 10.1073/pnas.0504557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chernoff YO, et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 38.Kushnirov VV, et al. Prion properties of the Sup35 protein of yeast Pichia methanolica. Embo J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayashiki T, et al. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell. 2001;7:1121–1130. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 40.Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 2009;5:e1000517. doi: 10.1371/journal.pgen.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giacomelli MG, et al. The conversion of 3′ UTRs into coding regions. Mol Biol Evol. 2007;24:457–464. doi: 10.1093/molbev/msl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green SR, Johnson AD. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:4191–4202. doi: 10.1091/mbc.E04-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hogan DJ, et al. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 45.Eaglestone SS, et al. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. Embo J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 48.Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2:135–140. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenthon JF, et al. The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot Cell. 2006;5:217–225. doi: 10.1128/EC.5.2.217-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senechal P, et al. The Schizosaccharomyces pombe Hsp104 disaggregase is unable to propagate the [PSI] prion. PLoS One. 2009;4:e6939. doi: 10.1371/journal.pone.0006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 53.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verstrepen KJ, Fink GR. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu Rev Genet. 2009;43:1–24. doi: 10.1146/annurev-genet-102108-134156. [DOI] [PubMed] [Google Scholar]
- 57.Bossdorf O, et al. Epigenetics for ecologists. Ecol Lett. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen JS, Otzen DE. Amyloid-a state in many guises: survival of the fittest fibril fold. Protein Sci. 2008;17:2–10. doi: 10.1110/ps.073127808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Bellot E, et al. The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. Embo J. 2000;19:3215–3222. doi: 10.1093/emboj/19.13.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derkatch IL, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? Embo J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santoso A, et al. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 62.Taneja V, et al. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edskes HK, et al. Nitrogen source and the retrograde signalling pathway affect detection, not generation, of the [URE3] prion. Yeast. 2006;23:833–840. doi: 10.1002/yea.1398. [DOI] [PubMed] [Google Scholar]
- 64.Brachmann A, et al. Reporter assay systems for [URE3] detection and analysis. Methods. 2006;39:35–42. doi: 10.1016/j.ymeth.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 65.Brachmann A, et al. Prion generation in vitro: amyloid of Ure2p is infectious. Embo J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexandrov IM, et al. Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation. J Biol Chem. 2008;283:15185–15192. doi: 10.1074/jbc.M802071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 68.Nakayashiki T, et al. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.King OD, Masel J. The evolution of bet-hedging adaptations to rare scenarios. Theor Popul Biol. 2007;72:560–575. doi: 10.1016/j.tpb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coustou V, et al. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunker AK, et al. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 72.Edskes HK, et al. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181:1159–1167. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beaumont HJ, et al. Experimental evolution of bet hedging. Nature. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- 74.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka M, et al. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 76.Cox B, et al. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satpute-Krishnan P, Serio TR. Prion protein remodelling confers an immediate phenotypic switch. Nature. 2005;437:262–265. doi: 10.1038/nature03981. [DOI] [PubMed] [Google Scholar]
- 78.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 79.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 80.Volkov KV, et al. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics. 2002;160:25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]