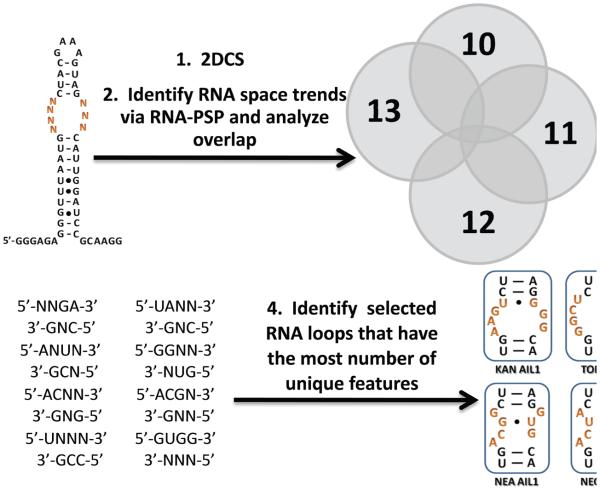
Figure 3.
The protocol utilized to choose selected RNA internal loops to study binding affinities and selectivities. The output of 2DCS is analyzed via the RNA-PSP program (18) to identify trends that are significant to a ≥95% confidence level. The trends for each ligand were then compared to identify those that are unique. Features that were specific for each aminoglycoside were then used to identify the selected internal loops that have the highest number of unique features. These were expected to be the most selective.