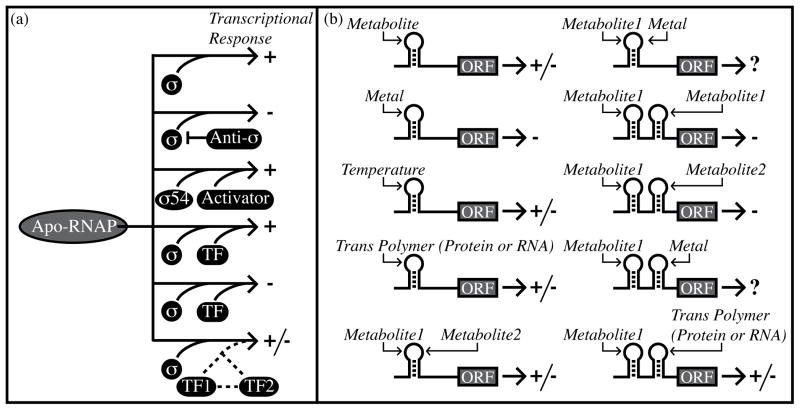
Figure 1.
Regulation of bacterial gene expression by transcriptional and posttranscriptional mechanisms. (a) Simplified view of transcriptional regulation using alternative sigma factors and DNA-binding transcription factors. Arrows point to the genetic outcome. Plus and minus signs indicate transcriptional activation and repression, respectively. Sigma factors promote transcription initiation through sequence-specific DNA contacts. Sigma 54 is an exception in that it requires a protein factor for transcriptional activation. Anti-sigma factors prevent sigma function. Transcription factors (“TF”) can individually activate or repress expression. Also, transcription factors can coordinate with one another to produce complex regulatory outcomes. (b) Simplified view of posttranscriptional regulation. Plus and minus symbols indicate increased or decreased gene expression. Hairpin structures denote individual cis-regulatory RNAs, which can respond to temperature, metabolites, metals, or trans-encoded polymers (RNAs or proteins). Alternatively, two different metabolites can associate with the same cis-regulatory RNA [25]. It is also possible that a particular, hypothetical cis-regulatory RNA may respond to a combination of metals and metabolite concentrations. Metabolite-sensing riboswitches may be arranged in tandem to respond to the same metabolite or to two different metabolite ligands [25,28]. It is also possible for multiple, distinct posttranscriptional regulatory mechanisms to cooperate in controlling downstream expression [30].