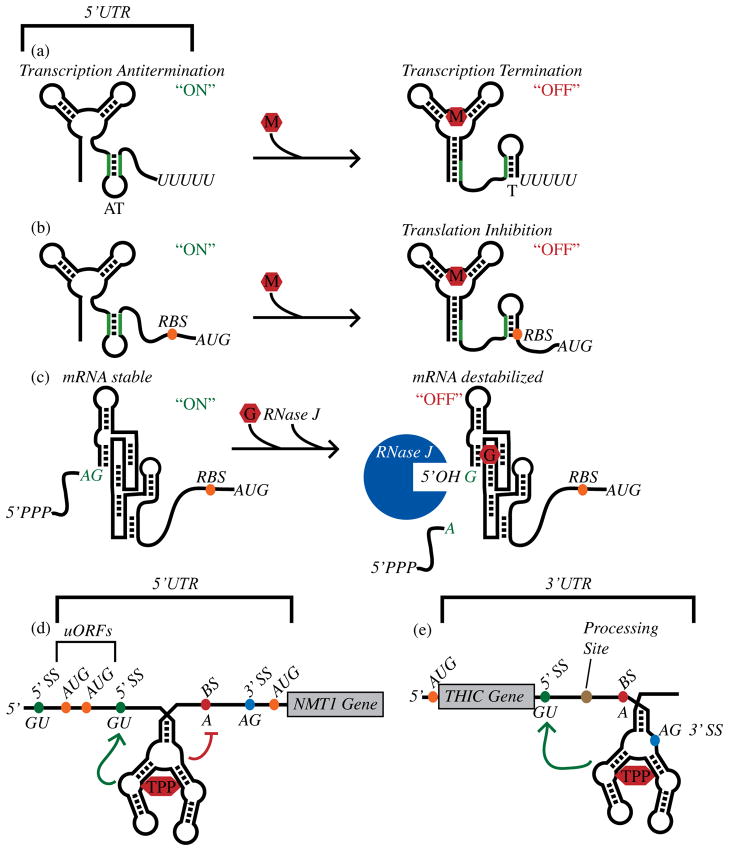
Figure 2.
Genetic control by metabolite-sensing riboswitches. (a) Control of formation of an intrinsic transcription terminator by a metabolite-binding riboswitch. “M” denotes the metabolite ligand. (b) Control of translation initiation efficiency by a metabolite-sensing riboswitch. (c) Control of mRNA stability by the GlcN6P-sensing ribozyme as described in the text. “G” denotes the GlcN6P riboswitch ligand. (d) An example of a TPP riboswitch that controls alternative splicing of the NMT1 gene in Neurospora crassa [37], as discussed in the text. “5′ SS” indicates the 5′ splice sites. “BS” indicates a branch site. (e) As discussed in the text, a TPP riboswitch from the 3′ UTR of Arabidopsis thaliana THIC controls mRNA processing and stability [38]. Green arrows signify regions where structural rearrangements occur upon TPP binding and that activate splicing elements. The red line identifies a region where structural rearrangements occur upon TPP binding and that repress splicing elements.