Abstract
Accurate measurement of cerebral lactate is critical to the understanding of brain function for psychiatric disorders such as panic disorder and bipolar disorder as well as mitochondrial dysfunction. Proton magnetic spectroscopic imaging (MRSI) techniques can be used to study lactate in vivo; however, accurate measurement of cerebral lactate, which is normally at low basal abundance, can be challenging. In this study, regional lactate measurements obtained with two different MRSI analytic approaches were evaluated using proton echo-planar spectroscopic imaging (PEPSI) data from 18 healthy adults participating in an in vivo sodium lactate infusion study. The results demonstrate that averaging data within a region of interest (ROI) before spectral fitting with LCModel results in significantly improved lactate measurement as compared to averaging chemical concentrations derived from the fitting of individual voxels in the ROI. Simulation results that confirm this finding are also presented. This study additionally outlines an atlas-based approach for the systematic computation of regional distributions of chemical concentrations in large MRSI data sets.
Keywords: magnetic resonance spectroscopy, brain bioenergetics, neuronal metabolism, panic disorder
1. Introduction
Cerebral lactate, a metabolic product of glycolysis, plays an integral role in neuronal energy metabolism (Schurr, 2006). Lactate exists in the healthy brain at low basal concentrations, and elevations can indicate transient changes in physiological state (van Rijen et al., 1989; Dager et al., 1999b; Friedman et al., 2007) or neural activation (Prichard et al., 1991; Sappey-Marinier et al., 1992; Frahm et al., 1996), as well as altered metabolic regulation such as in bipolar disorder (Dager et al., 2004) and panic disorder (Dager et al., 1994; Maddock, 2001). Other brain pathological states also exhibit characteristic brain lactate elevations, including tumors (Sijens et al., 1996), ischemia (Behar et al., 1983; Mathews et al., 1995), traumatic brain injury (Makoroff et al., 2005), and metabolic compromise from severe mitochondrial dysfunction, such as in MELAS (Kaufmann et al., 2004) or Leigh syndrome (Sijens et al., 2008). There is also increasing interest in brain lactate as a biomarker for less severe mitochondrial dysfunction (Lin et al., 2003)http://www.ninds.nih.gov/news_and_events/proceedings/20090629_mitochondrial.htm). Although proton magnetic resonance spectroscopy (MRS) provides a non-invasive means for lactate measurement in vivo, reliable quantification can be difficult, particularly at normal resting state. One known problem with measuring lactate and other chemicals at low natural abundance is systematic over-estimation when using linear combination fitting algorithms to estimate concentrations (Tkac et al., 2002; Kreis, 2004). A number of investigators have demonstrated that chemical estimation reliability can be increased when data signal-to-noise ratio (SNR) is improved by increasing the number of signal acquisitions or the static magnetic field strength (Tkac et al., 2002; Otazo et al., 2006; Posse et al., 2007).
At the same time, advances in magnetic resonance spectroscopic imaging (MRSI) have substantially increased the number of spectra that can be acquired in a given time. Two-dimensional proton MRSI techniques that can produce hundreds of usable spectra within a relatively short scan duration of 5–10 minutes are readily available on most clinical and research scanners, and three-dimensional MRSI techniques that can generate thousands of individual spectra in a single scanning session are rapidly coming into more common usage (Dager et al., 2008). Advances in MRSI data processing and analytic procedures have provided major improvements in the processing of large arrays of spectra (e.g. MIDAS (Maudsley et al., 2006), DSX (http://godzilla.kennedykrieger.org), and 3DiMRSI (http://mrs.cpmc.columbia.edu/3dicsi.html)); however, there is a persistent need for anatomically-specific results generated with minimal operator bias.
One strategy for summarizing the information in MRSI data sets that has been applied by investigators is to average chemical information across voxels within specific regions of interest (ROIs). This strategy preserves regionally specific information while also reducing the number of final calculated chemical concentrations to a manageable number. The predominantly used practice in carrying out this strategy is to first calculate individual voxel chemical concentration estimates and to then average these values across all voxels within an ROI. An alternative approach is to average data across voxels within the ROI prior to spectral fitting and chemical concentration estimation.
In this work we investigate whether averaging free induction decays (FIDs) prior to spectral fitting (referred to here as the spectral enhanced averaging method, or SEAM), instead of averaging chemical concentrations after fitting individual voxel data (referred to here as the individual voxel averaging method, or IVAM), can capitalize upon the benefits of improved spectral SNR to yield improved estimates of cerebral lactate when used in conjunction with fitting with LCModel (Provencher, 1993), a widely used linear combination fitting program for spectroscopic data. We analyze in vivo brain lactate data in healthy subjects at baseline levels, then longitudinally in response to intravenous sodium lactate infusion, utilizing ROIs defined through automated coregistration of proton echo-planar spectroscopic imaging (PEPSI) volumes to an anatomical atlas. As a point of reference, we also present findings for the measurement of NAA, which has a robust 1H MRSI signal. We integrate this technique of averaging across ROIs into a systematic method that can be applied to 2D and 3D MRSI data acquired at any field strength.
2. Methods
2.1 Subjects
MR data from 18 healthy control adults (8 males and 10 females ranging from 18 to 53 years of age) acquired as part of an intravenous sodium lactate infusion study of panic disorder, as previously described (Dager et al., 1994; Dager et al., 1999a), were included in this analysis. All subjects were medication free and fasting at the time of MRS evaluation, had no history of Axis I DSM-IV defined psychiatric disorders, and were not taking psychotropic medications. All subjects provided written consent for participation in the original study, which was approved by the University of Washington Human Subjects Review Committee.
2.2 Study procedures
All imaging data were acquired on a 1.5 Tesla GE Signa Horizon whole body MR scanner with version 5.8 Genesis operating software (Milwaukee, WI). A custom receive-only linear bird-cage radiofrequency head coil with a built-in head holder to immobilize the head, developed at the University of Washington, was used for all data collection.
High-resolution axial proton density and T2-weighted MR images were acquired using a fast spin echo pulse sequence with: TE=30/80 ms, TR= 3000 ms, FOV=220×220 mm, acquisition matrix=192×256 (reconstructed to 256×256), slice thickness=3 mm, gap=0 mm, number of slices=50. MRSI data were acquired using two-dimensional proton echo-planar spectroscopic imaging (PEPSI) (Posse et al., 1997) for an axial section of 20 mm in thickness at the level of the lateral ventricles. All PEPSI scans were acquired with a spatial matrix size of 32x32 voxels, 512 echo-planar echoes (sorted to 256 odd echoes and 256 even echoes) and a 22 cm FOV. An initial baseline water-suppressed (“metabolite”) scan with a TE of 20 ms was acquired, followed by a baseline non-water-suppressed (“water”) scan with a TE of 20 ms. Ten metabolite scans [TR=2000 ms, TE=144 ms] were then serially acquired. The TE of 144 ms was chosen to invert the lactate peak to minimize effects of overlapping lipid signal on lactate fitting during spectral processing. Three metabolite scans, acquired over 15 minutes at baseline, were followed by four scans during intravenous infusion of 0.5 mol/L sodium lactate (10 ml/kg total) over 20 minutes, and three post-infusion scans during a 15 minute recovery period. Two of the subjects were able to complete only 9 out of 10 MRSI scans, and one subject completed only 8 of the scans. Data from the initial metabolite scan with a TE of 20 ms are not described in the current analysis.
2.3 Structural image segmentation
Proton density (PD)-weighted and T2-weighted images were segmented to produce a cerebral spinal fluid (CSF) image for each subject. To do this, each PD and T2 weighted image at a level corresponding to the location of the PEPSI slab was first corrected for RF inhomogeneity using homomorphic filtering (Guillemaud, 1998). Corrected images were then classified using a k-means algorithm to produce binary CSF images and averaged together. The resulting image was filtered with a 3 pixel wide Gaussian kernel to produce a final CSF image with a point spread function approximating that of the spectroscopic images (Friedman et al., 2003). The CSF images were used in partial volume correction during chemical quantification.
2.4 Spectroscopic data analysis
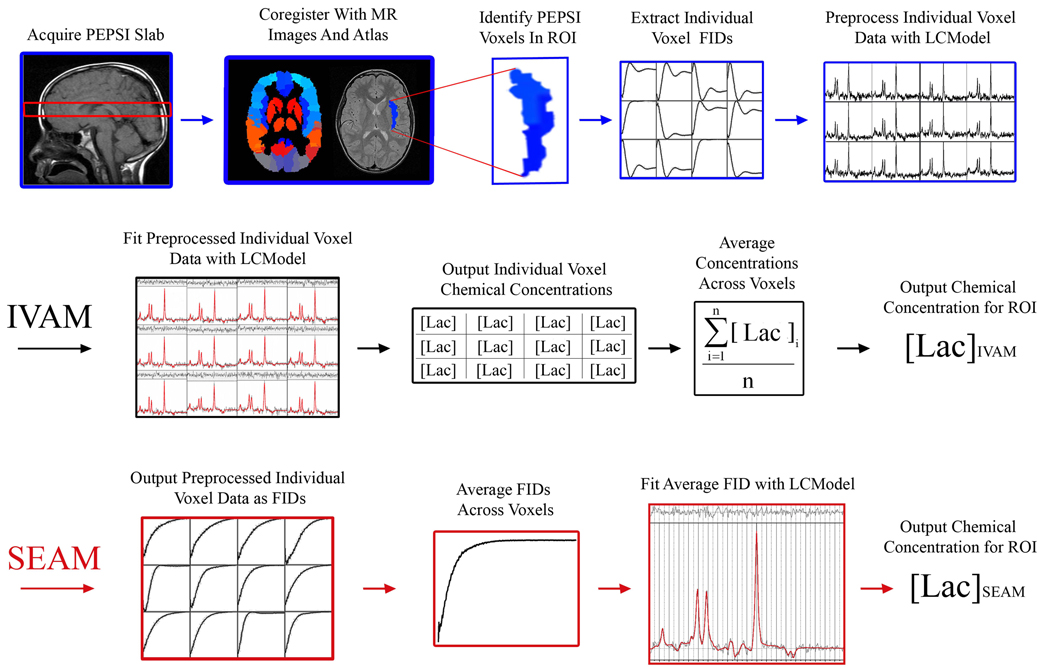
A schematic representation of the systematic approach for analysis of MRSI data is shown in Figure 1. Data pre-processing steps are shown in the top row. The bottom two rows outline the steps used for the IVAM and SEAM approaches, which are detailed below.
Figure 1.
Schematic representation of the systematic analysis of MRSI data used in this study. Top row: Data pre-processing steps. Middle row: Steps for computing chemical concentration estimates with IVAM. Bottom row: Steps for computing chemical concentration estimate with SEAM. In this figure, lactate is used as an example chemical of interest, the right insula is used as an example region of interest, and all spectroscopic data shown are from the sodium lactate infusion study.
2.4.1 Raw data reconstruction and co-registration with atlas
Spectroscopic data were reconstructed using custom software developed in our laboratory. A Gaussian filter was applied in both spatial dimensions to the raw PEPSI data to minimize the Gibbs artifact. The data were then Fourier transformed to produce a three-dimensional matrix with two spatial dimensions and a chemical shift dimension. PEPSI slabs were coregistered to the T2 images for each subject using spatial information from the image file headers. The T2 images were then coregistered to the Anatomic Labeling Map (AAL) anatomical atlas (http://www.sph.sc.edu/comd/rorden/mricro.html) using the FLIRT toolbox of the FSL software (http://www.fmrib.ox.ac.uk/fsl/index.html, 12 affine transform). This whole brain atlas divides the brain into 116 brain regions based on anatomic gyral structure. Twenty-two ROIs of varying size were defined using this atlas. The T2 images and coregistered atlas were used to assign anatomical locations to each PEPSI voxel. After this coregistration was complete, the PEPSI spectra were inverse Fourier transformed and saved to a file for input into the LCModel software package (v.6.2) (Provencher, 1993).
2.4.2 Individual voxel correction and spectral fitting
LCModel was first used to perform peak alignment and zero-order phase correction on all data from both the water and metabolite scans. For the IVAM method, spectral fitting was additionally performed at this stage, and the resulting concentration estimates, as well as associated Cramer-Rao lower bounds (CRLBs) for each echo, were output to a file. All fitting with LCModel was performed using a simulated spin-echo basis set. LCModel “.coraw” output files containing the inverse Fourier transform (or FIDs) of both chemical and water spectra after LCModel peak alignment and phase correction were generated for use with the SEAM averaging method.
2.4.3 Identification and exclusion of low-quality spectra
A custom in-house interactive program with a graphical user interface was developed using MATLAB (v. 7.5, The Mathworks, Inc) to aid in overall quality analysis of the spectra for each subject, and determination of appropriate thresholds for exclusion of poor quality spectra. These evaluations were performed based on output generated by the initial LCModel processing. For each subject, the visualization tool displayed overlays of spectra that would be included and excluded from analysis, according to editable criteria, along with a display of the location of included voxels on the PEPSI slab. The tool also allowed for sequential viewing of individual spectra, along with associated LCModel output values, including full width at half maximum (FWHM) and chemical CRLBs, which are widely used in MR spectroscopy as a measure of the relative uncertainty, or precision, of calculated concentration estimates (Cavassila et al., 2001; Jiru et al., 2006). For this study, an individual voxel spectrum was excluded if the initial LCModel fit calculated a CRLB greater than or equal to a value of 20% for NAA, if the LCModel FWHM value exceeded 0.1 ppm, or if the water concentration in a voxel was less than five standard deviations of the mean of the water concentration in a 10×10 voxel region centered in the middle of the PEPSI slab.
2.4.4 Averaging of voxel data using IVAM
LCModel values for each individual voxel in the ROI were converted to chemical concentrations according to the quantification method detailed below. For the water correction, each voxel chemical concentration estimate was corrected by the water concentration estimate for the same voxel. After quantification, chemical concentrations were averaged across voxels in each of the twenty-two brain regions.
2.4.5 Averaging of voxel data using SEAM
For both the water and chemical data, time domain phased and peak-aligned FIDs generated by the LCModel program were averaged for all voxels in each region to produce a single FID for the region. This averaged FID was fit using LCModel with the same settings used for the individual voxel fitting. LCModel values were converted to chemical concentrations according to the quantification method detailed below.
2.4.6 Chemical quantification
Chemical concentrations were computed using methods detailed in the LCModel User's Manual (available at http://s-provencher.com/pages/lcm-manual.shtml), and following procedures used in our past work (Friedman et al., 2003; Friedman et al., 2006). After line-fitting, corrections were made to the LCModel output using software developed in-house with Matlab. Chemical and water amplitudes were corrected for acquisition parameters (receiver settings, transmitter gain) and voxel size. Water amplitudes were adjusted for water molarity (using an estimated value of 35900 mM, which is intermediate between the molarity of water in gray and white matter) and attenuation of the water signal due to relaxation effects and other effects that affect only the unsuppressed water reference signal (estimated as 0.7) (LCModel User's Manual). These adjusted amplitudes were then multiplied by the tissue fraction within the voxel as determined from the segmented CSF maps. The water amplitudes were corrected for relaxation, with values derived from the literature [T1=515 ms; T2=85 ms (Barker et al., 1993)]. Following these corrections, the chemical amplitude for each voxel was referenced to the water amplitude based on internal water referencing techniques (Barker et al., 1993). The concentrations of NAA and lactate were then corrected for T1 and T2 relaxation using literature values [NAA: T1=1450 ms, T2=450 ms; lactate: T1=1550 ms, T2=1200 ms (Frahm et al., 1989)].
2.5 Statistical methods
Statistical analyses of in vivo findings for selected ROIs were performed with SPSS (v.14.0) (Chicago, IL) and Matlab. The three ROIs chosen to illustrate differences in the analytic approaches were: a right frontal lobe region, which is an area where MRS data is usually difficult to measure due to susceptibility artifacts; a left insular region, which is of particular interest in panic disorder; and the PEPSI volume as a whole. Repeated measures analysis of variance (ANOVA), with Bonferroni correction, was used to test for the main effects of spectral analytic approach on estimated chemical concentrations, collapsed over all time points of the infusion experiment. For ANOVAs reaching significance (P<0.05), post-hoc two-tailed paired t-tests, assuming equal variances between groups, were performed to test for differences between the mean concentration values produced by the two methods at individual time points.
2.6 Simulation
Comparisons of the effect of additive noise on the lactate concentration estimates produced with the SEAM and IVAM approaches were evaluated in two simulation experiments as detailed below. For both simulation experiments, SNR values were calculated in the Fourier domain by dividing the amplitude of the NAA peak by twice the standard deviation of a segment of the spectrum with no signal.
2.6.1 Effect of noise on lactate detection - no originally detected lactate signal
Simulated random noise at a range of amplitudes was added to an FID derived from a single subject’s data. The FID was calculated by averaging all of the FIDs from all valid voxels of the PEPSI slab from the subject at one time point during the baseline period prior to lactate infusion. The LCModel estimated lactate concentration for this averaged FID, before the addition of noise, was equal to zero. Noise was added to both the real and imaginary parts of the FID.
2.6.2 Effect of noise on lactate detection - lactate signal originally present
Fifty simulated voxel FIDs were generated by adding normally distributed random noise to the real and imaginary components of an FID that was created by averaging the FIDs for all valid voxels at the last time point in the infusion period for all study subjects. SEAM and IVAM were used to calculate lactate concentrations for simulated ROIs consisting from one to fifty of these simulated voxels. This experiment was performed for ten different random noise sets, with the same noise amplitude across sets, and the means and standard deviations of the concentration values computed by the two methods across all experiments were calculated. For the SEAM method, mean CRLBs across all experiments were additionally computed for each ROI size.
3. Results
In the identification of usable spectra from the in vivo sodium lactate infusion experiment, the NAA CRLB threshold alone resulted in the inclusion of 96% of voxels in the left insular region, 60% of the voxels in the right frontal lobe region, and 61% of the voxels in the whole slab ROI. Addition of the FWHM and water signal quality threshold resulted in a much more conservative number of included voxels, with 63% of the spectra included for the left insular region, 27% of the spectra included in the right frontal region, and 27% of spectra included in the whole slab ROI. Across subjects, the average numbers of voxels included in each ROI at baseline were 36 voxels for the left insular region, 52 voxels for the right frontal region, and 229 voxels for the whole slab ROI. The additional conservative criterion that each voxel is valid at all time points due to the longitudinal nature of this study reduced the average number of voxels in the ROIs to 21 for the left insular region, 30 for the right frontal region, and 131 for the whole slab ROI. The results presented below correspond to these latter, more conservative, ROI voxel numbers.
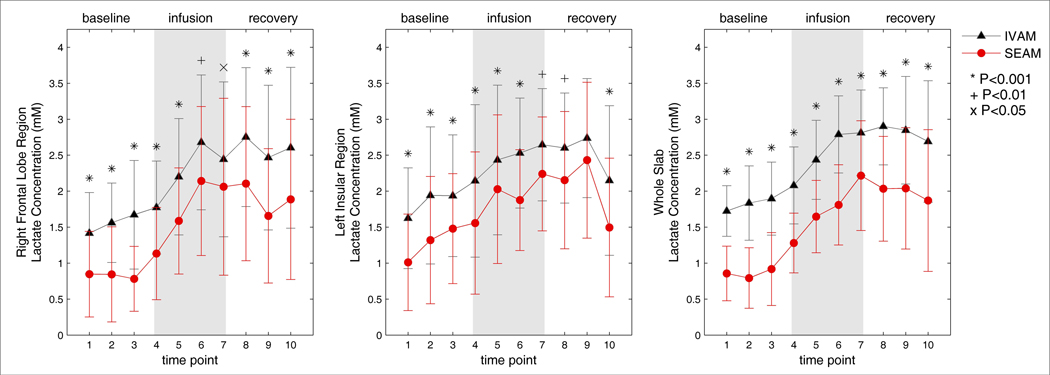
In vivo concentration estimates for lactate calculated with IVAM and SEAM for the right frontal lobe region, the left insular region, and the PEPSI slab as a whole, averaged across all subjects, are shown in Figures 2. Error bars indicate the standard deviation across subjects and, if significant differences were found across all time points, additional markers at each time point indicate the level of significance. All three regions showed the expected general progressive rise of lactate concentration during the infusion period. The lactate concentration estimates for all three regions were found to be significantly reduced for SEAM as compared to IVAM (all F > 4.6; P< 0.041).
Figure 2.
Concentration estimates for lactate for the IVAM and SEAM methods over the duration of the dynamic infusion experiment for the right frontal lobe region (left), left insular region (middle) and whole slab (right). Mean concentrations over all 18 subjects are shown with error bars indicating the standard deviation. Additional markers denote the level of significance of differences in the means at each time point.
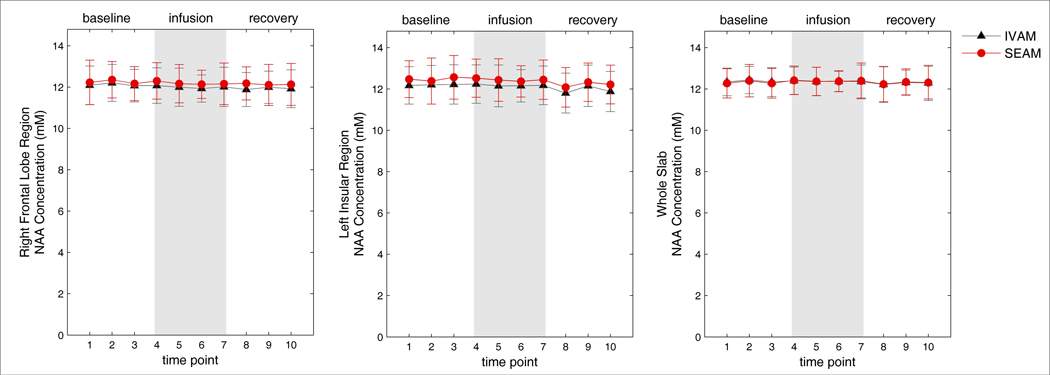
In vivo concentration estimates for NAA calculated by the two analytic approaches for the three ROIs are shown in Figure 3. NAA concentration estimates remained relatively consistent across time points for all regions, as expected, for both IVAM and SEAM, and the repeated measures ANOVA analyses found no significant differences in concentration estimates produced by the two analytic approaches in any of the three regions (all F <0.799; P> 0.380).
Figure 3.
Concentration estimates for NAA for the IVAM and SEAM methods over the duration of the dynamic infusion experiment for the right frontal lobe region (left), left insular region (middle) and whole slab (right). Mean concentrations over all 18 subjects are shown with error bars indicating the standard deviation across subjects.
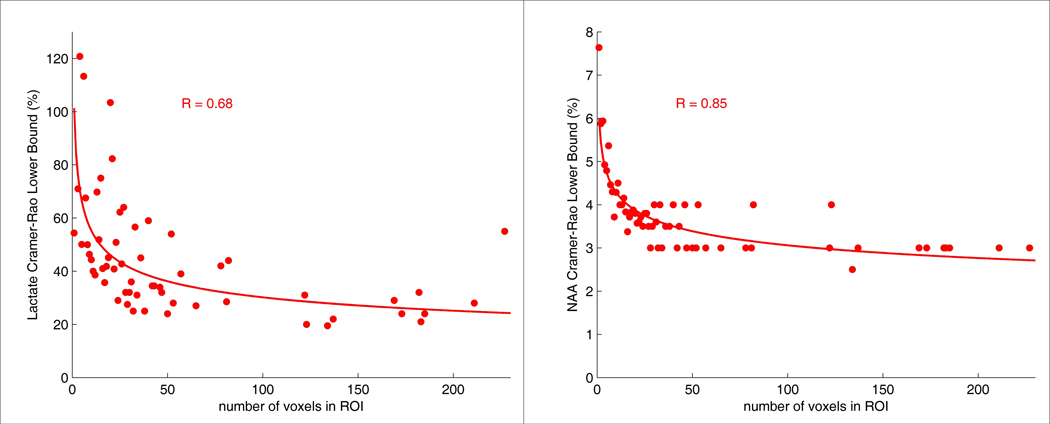
The effect of the size of the ROI on the CRLBs with SEAM for the in vivo data is shown in Figure 4. These plots represent the results from analyzing all 22 ROIs across subjects and show the average CRLB for each ROI size. Trend lines have been added to illustrate approximate functional relationships between the number of voxels in the ROI and the CRLBs. These plots indicate that the average CRLBs produced by SEAM decreased for both chemicals as the size of the ROI increased.
Figure 4.
Average CRLBs across all subjects and all twenty-two brain regions plotted as a function of the number of voxels in the ROI for cerebral lactate (left) and NAA (right) for the SEAM approach. The data corresponds to the last time point of the lactate infusion period. The equations for the SEAM lactate and NAA trend lines are y=101.32*x−0.26 and y=6.00*x−0.15, respectively.
An example of the potential effects of additive simulated random noise on lactate quantification with LCModel for a spectrum with no lactate signal is shown in Figure 5. The left panel of this figure shows the spectrum with no added simulated noise (gray waveform), with the LCModel fit superimposed (red waveform). The fit of this spectrum yielded an estimated lactate concentration of zero. The middle and right panels show the effects of two cases of adding simulated noise to this spectrum where the LCModel algorithm interpreted random fluctuations as lactate peaks. Although the original spectrum contained no lactate signal, addition of noise created an apparent lactate signal detected by LCModel. Further, as the SNR decreased due to increased noise, the concentration of lactate detected by LCModel increased.
Figure 5.
Effect of adding simulated noise to in vivo spectroscopy data on quantification of brain lactate by LCModel. The in vivo spectrum is represented in gray, and red lines indicate the LCModel fit. Left: The original spectrum, calculated as the average across all voxels in the PEPSI slab for a single subject, demonstrates no lactate signal detected by LCModel (a meaningless CRLB of 999% is produced when LCModel can not fit the chemical of interest). Middle and right: The same spectrum with two different amplitudes of noise added. The LCModel fits detected noise as lactate signal for both of these spectra.
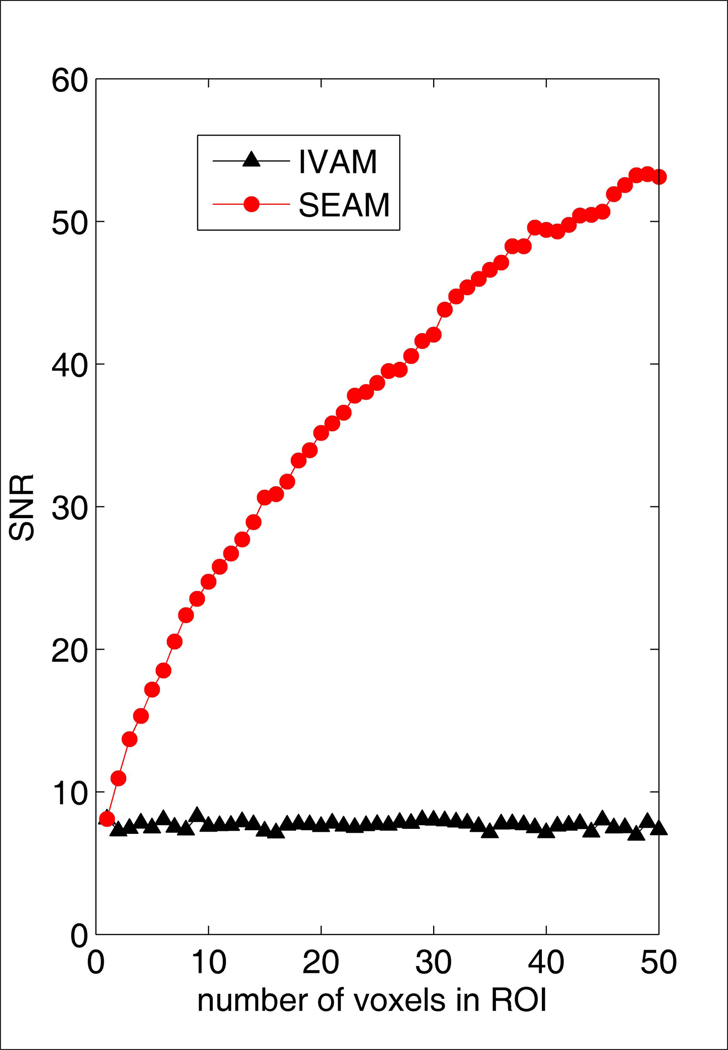
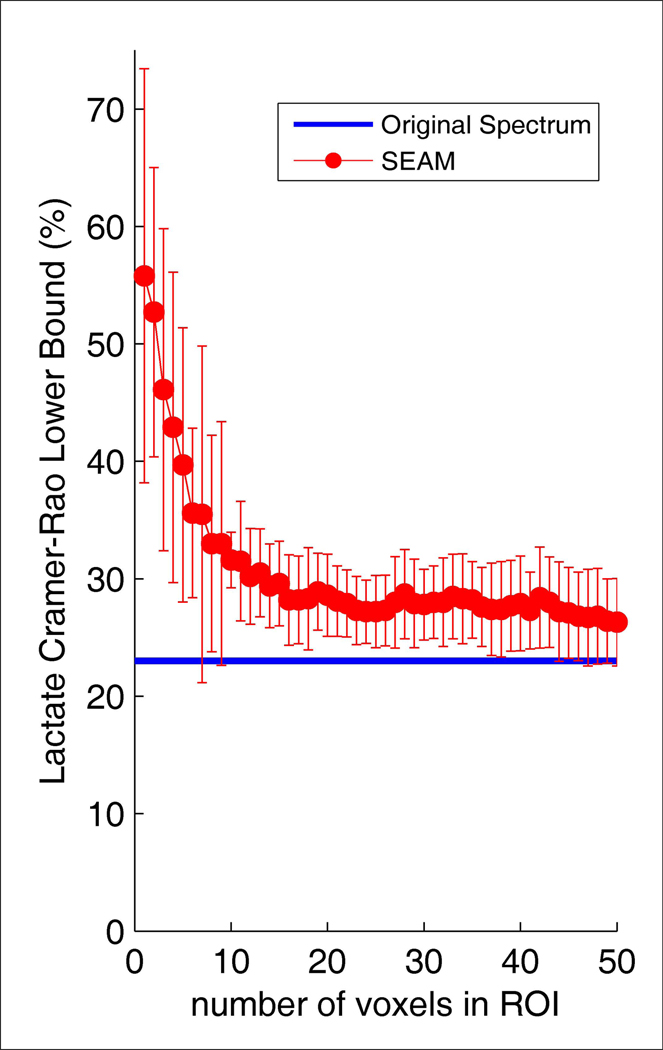
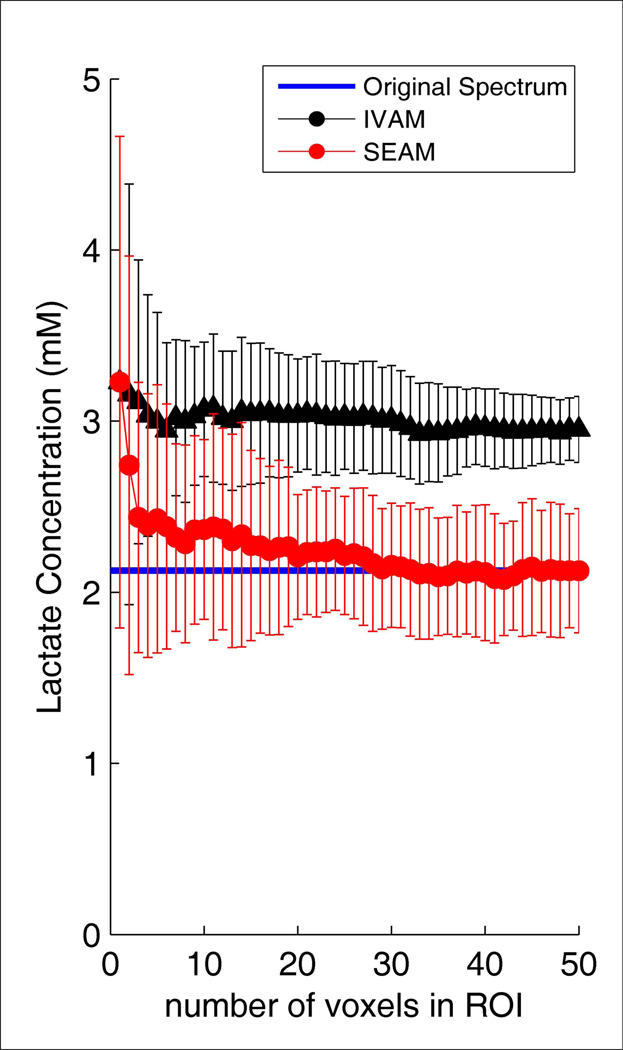
Simulation results from adding random noise to a spectrum computed by averaging all valid voxel data at the last time point during infusion across all subjects are shown in Figure 6– Figure 8. The SNR values in Figure 6 for SEAM were calculated from the average spectrum in the simulated ROI. The SNR values for IVAM were calculated as the average SNR across all individual voxels in the ROI. This figure demonstrates that while increasing the size of the ROI resulted in improved SNR of the fitted spectrum for SEAM (as expected, the SNR for the SEAM approach was proportional to the square root of the number of voxels averaged (r= 0.999)), the size of the ROI had no effect on the SNR of the fitted spectra with the IVAM approach. The effect of increasing the number of voxels in the simulated ROI on lactate concentration estimates is shown in Figure 7. The blue line indicates the values for the original spectrum with no added noise. As the number of voxels increased, the SEAM concentration estimates converged to a stable value that closely approximated the original spectrum lactate concentration. Conversely, IVAM yielded lactate concentration values that were consistently elevated for all ROI sizes compared to both the SEAM and the original spectrum lactate estimates. The variance in SEAM and IVAM values indicated by the standard deviation error bars demonstrate that the results generated by both methods were dependent upon background noise characteristics. Figure 8 shows that the SEAM CRLBs decreased progressively with increased ROI size and approached the lactate CRLB for the fitting of the original spectrum.
Figure 6.
SNR as a function of the number of voxels in simulated ROIs for the SEAM and IVAM approaches. Individual voxel data were calculated by adding simulated noise to a single FID created by averaging all valid FIDs at the last time point during the infusion period across all subjects. The mean SNR across ten experiments, each with a different random noise introduction, is shown. The SNR for the SEAM method is proportional to the square root of the number of voxels (r=0.999).
Figure 8.
LCModel CRLBs for the fitting of lactate as a function of number of voxels in the simulated ROIs for the SEAM analytic approach. The plots show the means and standard deviations across ten experiments, each with a different random noise introduction. The blue line indicates the value for the original spectrum.
Figure 7.
Lactate concentration estimates as a function of number of voxels in the simulated ROIs for the SEAM and IVAM methods, corresponding to the data presented in Figure 6. The plots show the means and standard deviations across ten experiments, each with a different random noise introduction. A blue line on each plot indicates the value for the original spectrum.
4. Discussion
Systematic handling of the large volumes on data contained in MRSI data sets can be achieved through automated coregistration to an anatomic atlas, and averaging of data within ROIs. Results from this study demonstrate that the approach used for combining data in ROIs can have a substantial effect on cerebral lactate concentration estimates. The sodium lactate infusion data set provided a valuable opportunity to investigate the effect of two different data averaging techniques on lactate measurement in the healthy adult brain, where robust detection was possible due to elevated lactate levels that would otherwise only occur under pathological conditions. With both the SEAM and IVAM analyses, lactate estimates demonstrated a distinct and progressive rise during the infusion. However, the SEAM estimates for lactate concentration were significantly decreased as compared to the IVAM estimates. Simulation showed that SEAM resulted in more accurate concentration values than IVAM, especially for large ROIs. The observed systematic over-estimation of lactate when fitting individual voxel data with IVAM is consistent with previous reports (Tkac et al., 2002; Kreis, 2004). As demonstrated through simulation, this over-estimation of concentration is likely the result of spurious detection of lactate by the LCModel fitting algorithm. Systematic over-estimation of concentration was not observed for NAA, likely because this chemical is not at low concentrations in the healthy human brain. It is important to note that the differences between the lactate concentration estimates produced by the IVAM and SEAM approaches are due to a bias introduced by the LCModel algorithm when fitting signals with low SNR, and are not attributable to the analytic techniques themselves. Indeed, if LCModel never identified noise as lactate signal, for large ROIs, the IVAM and SEAM techniques would produce more similar values. The results from this study additionally demonstrate that increasing the size of the ROI, and hence the number of measurements, does not eliminate lactate concentration estimate differences between the two approaches.
Previous studies have investigated the effect of SNR on LCModel quantification accuracy for single voxel MRS data acquired at short echo times using simulated (Kanowski et al., 2004; Macri et al., 2004), and in vivo data (Tkac et al., 2002). Reproducibility of LCModel quantification of MRSI data has also been assessed in a number of studies (Jackson et al., 1994; Tedeschi et al., 1995; Tedeschi et al., 1996; Li et al., 2002; Gu et al., 2008). To our knowledge, the accuracy of LCModel quantification of cerebral lactate and the benefits and pitfalls of different methods for signal averaging have not been previously addressed. Given that lactate exists at low basal concentrations and is of interest in a variety of neuroscience and psychiatric applications, the ability to utilize the large volume of MRSI data to increase the accuracy in the measurement of this chemical is extremely valuable.
In addition to the improved accuracy in the measurement of lactate with SEAM, the results from this study demonstrate that for this analytic approach, the CRLBs, and thus the estimate reliability for both lactate and NAA, improve as the number of voxels in the ROI (and hence SNR of the averaged spectrum) increases. For IVAM, the reliability of the chemical estimate is also expected to improve as the number of voxels in the ROI increases, due to the increased number of measurements, but this is not reflected in the CRLBs for the individual voxel fits.
One important corollary of the findings for SEAM is that with this analytic approach, concentration differences observed between ROIs in the same brain, and across individuals, may reflect differences in SNR (caused, for example, by number of constituent voxels) rather than differences in actual chemical content. As noted by other investigators (Kreis, 2003), this may be best controlled for by ensuring that the SNR of spectra are similar when making comparisons of chemical concentration estimates within or across subjects. It is also important to keep in mind that the benefits demonstrated here with SEAM assume a simplified view that a brain region has a homogeneous distribution of the chemical of interest, with SNR as the primary measurement limitation. SEAM does result in a loss of intra-regional information, and a hybrid IVAM/SEAM approach for measurement of high SNR chemicals may be useful to maintain measures of variability within ROIs. Issues of differential gray and white matter relaxation properties and partial volume effect for lactate quantification are important considerations (Friedman et al., 2006) but beyond the scope of this report. Additionally, the simulation findings presented in this study are meant as a proof of principle to demonstrate the effects of two analytic methods on spectra with added random noise. More extensive simulation would be necessary for a more thorough characterization of the behavior of LCModel fitting for data with different noise characteristics in order to quantify the benefits of SEAM over IVAM under varied conditions.
The systematic analysis approach presented here facilitates the processing of MRSI data across brain regions. Steps that ease processing include the use of uniform thresholds for spectral quality across all study subjects, and coregistration of an anatomic atlas to individual subject anatomy in order to automate ROI determination. It is important to note, however, that even with this level of automation, careful attention to the results of every step taken during MRSI data analysis is critical, as signal characteristics and artifacts can differ greatly across studies due to the many factors involved in the acquisition process. In particular, we found that during preprocessing, careful inspection of individual subject data using our graphical user interface visualization tool was essential to ensure that the criteria used to detect and remove low quality spectra were appropriate for all study data. The visualization tool described here provided a relatively fast and simple way to view the data for every subject and evaluate spectral quality in a comprehensive and efficient manner.
The approach outlined here that can be applied to a wide range of applications. ROI selection can be custom tailored to address specific research or clinical questions and the procedure used can have a large effect on the quantification. Future work can address in more detail optimal approaches for spatial localization within ROIs, which will particularly require additional refinement for application to three-dimensional MRSI data sets.
In our analyses, we did not exclude spectra with lactate CRLBs above a specific threshold, as is a relatively common practice in the literature since CRLBs are linked to chemical estimation reliability. However, due to the difficulty in discriminating lactate signal from background noise, the meaning of the CRLBs produced by LCModel for this chemical is less clear. In analysis of these data, we observed that large spurious lactate concentrations are often associated with low CRLBs. Consistent with our observations, others have noted that CRLBs produced by LCModel can be artificially low for estimation of chemicals at low abundance (Kreis, 2004). Further, a high CRLB can result from the absence of lactate signal in a voxel, which can lead to inappropriate spectral rejection. Discarding spectra for which no lactate is detectable biases concentration estimates towards higher values. In the absence of clear standards for deciding whether a lactate concentration estimate should be included in reported results based on lactate CRLBs, deriving criteria from other measures of overall spectral quality may avoid this complication. Analytic approaches, such as SEAM, that improve the SNR of fitted spectra, can provide a more realistic assessment of brain lactate levels than evaluating lactate concentrations based on measures derived from the fitting of individual spectra.
This study has demonstrated the benefits of SEAM in improving lactate measurement for data acquired at 1.5T. Higher field strength scanners, most typically 3T, are now coming into more common usage in clinical and research settings, and can significantly increase the SNR of the lactate signal. Increased SNR can make the lactate signal easier to discriminate from background noise by the LCModel fitting algorithm, which could potentially reduce the benefits of SEAM as compared to IVAM in estimating lactate concentrations. However, current progress in MRSI is highly focused on capitalizing upon the higher SNR provided by high field strengths to increase brain coverage, while at the same time reducing voxel sizes and acquisition times. As there is a tradeoff between SNR, spatial resolution, and scan time, these adjustments are likely to cause individual voxel lactate signal SNR to remain relatively low for many studies, resulting in similar overestimation issues when fitting with LCModel, as demonstrated in this report. For this reason, the SEAM analytic approach will likely continue to be valuable in the analysis of MRSI data acquired at higher fields.
In summary, we present a systematic approach for the analysis of MRSI data sets based on coregistration of spectroscopy data with an anatomical atlas and the use of SEAM, a method that capitalizes on the large quantity of information contained in MRSI data sets to improve lactate concentration estimation accuracy for regions of interest. These improvements are achieved in the data processing stage, and can augment the benefits of improving data quality at the level of acquisition, such as by increasing the static magnetic field strength. This systematic approach will be increasingly valuable as MRSI acquisition techniques, and the amount of spectral data they generate, continue to progress.
Acknowledgements
This study was supported by National Institutes of Health grants RO1-2MH50579 and 1P50HD55782.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker PB, Soher BJ, Blackband SJ, Chatham JC, Mathews VP, Bryan RN. Quantitation of proton NMR spectra of the human brain using tissue water as an internal concentration reference. NMR in Biomedicine. 1993;6:89–94. doi: 10.1002/nbm.1940060114. [DOI] [PubMed] [Google Scholar]
- Behar KL, den Hollander JA, Stromski ME, Ogino T, Shulman RG, Petroff OA, Prichard JW. High-resolution 1H nuclear magnetic resonance study of cerebral hypoxia in vivo; Proceedings of the National Academy of Sciences of the United States of America; 1983. pp. 4945–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramer-Rao bounds: an evaluation tool for quantitation. NMR in Biomedicine. 2001;14:278–283. doi: 10.1002/nbm.701. [DOI] [PubMed] [Google Scholar]
- Dager SR, Corrigan NM, Richards TL, Posse S. Research applications of magnetic resonance spectroscopy to investigate psychiatric disorders. Topics in Magnetic Resonance Imaging. 2008;19:81–96. doi: 10.1097/RMR.0b013e318181e0be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dager SR, Friedman SD, Heide A, Layton ME, Richards T, Artru A, Strauss W, Hayes C, Posse S. Two-dimensional proton echo-planar spectroscopic imaging of brain metabolic changes during lactate-induced panic. Archives of General Psychiatry. 1999a;56:70–77. doi: 10.1001/archpsyc.56.1.70. [DOI] [PubMed] [Google Scholar]
- Dager SR, Friedman SD, Parow A, Demopulos C, Stoll AL, Lyoo IK, Dunner DL, Renshaw PF. Brain metabolic alterations in medication-free patients with bipolar disorder. Archives of General Psychiatry. 2004;61:450–458. doi: 10.1001/archpsyc.61.5.450. [DOI] [PubMed] [Google Scholar]
- Dager SR, Layton ME, Strauss W, Richards TL, Heide A, Friedman SD, Artru AA, Hayes CE, Posse S. Human brain metabolic response to caffeine and the effects of tolerance. American Journal of Psychiatry. 1999b;156:229–237. doi: 10.1176/ajp.156.2.229. [DOI] [PubMed] [Google Scholar]
- Dager SR, Marro KI, Richards TL, Metzger GD. Preliminary application of magnetic resonance spectroscopy to investigate lactate-induced panic. American Journal of Psychiatry. 1994;151:57–63. doi: 10.1176/ajp.151.1.57. [DOI] [PubMed] [Google Scholar]
- Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hanicke W, Sauter R. Localized proton NMR spectroscopy in different regions of the human brain in vivo. Relaxation times and concentrations of cerebral metabolites. Magnetic Resonance in Medicine. 1989;11:47–63. doi: 10.1002/mrm.1910110105. [DOI] [PubMed] [Google Scholar]
- Frahm J, Kruger G, Merboldt KD, Kleinschmidt A. Dynamic uncoupling and recoupling of perfusion and oxidative metabolism during focal brain activation in man. Magnetic Resonance in Medicine. 1996;35:143–148. doi: 10.1002/mrm.1910350202. [DOI] [PubMed] [Google Scholar]
- Friedman SD, Jensen JE, Frederick BB, Artru AA, Renshaw PF, Dager SR. Brain changes to hypocapnia using rapidly interleaved phosphorus-proton magnetic resonance spectroscopy at 4 T. Journal of Cerebral Blood Flow and Metabolism. 2007;27:646–653. doi: 10.1038/sj.jcbfm.9600383. [DOI] [PubMed] [Google Scholar]
- Friedman SD, Shaw DW, Artru AA, Dawson G, Petropoulos H, Dager SR. Gray and white matter brain chemistry in young children with autism. Archives of General Psychiatry. 2006;63:786–794. doi: 10.1001/archpsyc.63.7.786. [DOI] [PubMed] [Google Scholar]
- Friedman SD, Shaw DW, Artru AA, Richards TL, Gardner J, Dawson G, Posse S, Dager SR. Regional brain chemical alterations in young children with autism spectrum disorder. Neurology. 2003;60:100–107. doi: 10.1212/wnl.60.1.100. [DOI] [PubMed] [Google Scholar]
- Gu M, Kim DH, Mayer D, Sullivan EV, Pfefferbaum A, Spielman DM. Reproducibility study of whole-brain 1H spectroscopic imaging with automated quantification. Magnetic Resonance in Medicine. 2008;60:542–547. doi: 10.1002/mrm.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemaud R. Uniformity correction with homomorphic filtering on region of interest. IEEE ICIP. 1998;2:872–875. [Google Scholar]
- Jackson EF, Doyle TJ, Wolinsky JS, Narayana PA. Short TE hydrogen-1 spectroscopic MR imaging of normal human brain: reproducibility studies. Journal of Magnetic Resonance Imaging. 1994;4:545–551. doi: 10.1002/jmri.1880040406. [DOI] [PubMed] [Google Scholar]
- Jiru F, Skoch A, Klose U, Grodd W, Hajek M. Error images for spectroscopic imaging by LCModel using Cramer-Rao bounds. MAGMA. 2006;19:1–14. doi: 10.1007/s10334-005-0018-7. [DOI] [PubMed] [Google Scholar]
- Kanowski M, Kaufmann J, Braun J, Bernarding J, Tempelmann C. Quantitation of simulated short echo time 1H human brain spectra by LCModel and AMARES. Magnetic Resonance in Medicine. 2004;51:904–912. doi: 10.1002/mrm.20063. [DOI] [PubMed] [Google Scholar]
- Kaufmann P, Shungu DC, Sano MC, Jhung S, Engelstad K, Mitsis E, Mao X, Shanske S, Hirano M, DiMauro S, De Vivo DC. Cerebral lactic acidosis correlates with neurological impairment in MELAS. Neurology. 2004;62:1297–1302. doi: 10.1212/01.wnl.0000120557.83907.a8. [DOI] [PubMed] [Google Scholar]
- Kreis R. Issues of spectral quality in clinical 1H-magnetic resonance spectroscopy and a gallery of artifacts. NMR in Biomedicine. 2004;17:361–381. doi: 10.1002/nbm.891. [DOI] [PubMed] [Google Scholar]
- Kreis R, Boesch C. Bad spectra can be better than good spectra; Proceedings of the 11th Meeting of the International Society of Magnetic Resonance in Medicine; Toronto, USA. 2003. p. 264. [Google Scholar]
- Li BS, Babb JS, Soher BJ, Maudsley AA, Gonen O. Reproducibility of 3D proton spectroscopy in the human brain. Magnetic Resonance in Medicine. 2002;47:439–446. doi: 10.1002/mrm.10081. [DOI] [PubMed] [Google Scholar]
- Lin DD, Crawford TO, Barker PB. Proton MR spectroscopy in the diagnostic evaluation of suspected mitochondrial disease. AJNR. American Journal of Neuroradiology. 2003;24:33–41. [PMC free article] [PubMed] [Google Scholar]
- Macri MA, Garreffa G, Giove F, Guardati M, Ambrosini A, Colonnese C, Maraviglia B. In vivo quantitative 1H MRS of cerebellum and evaluation of quantitation reproducibility by simulation of different levels of noise and spectral resolution. Magnetic Resonance Imaging. 2004;22:1385–1393. doi: 10.1016/j.mri.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Maddock RJ. The lactic acid response to alkalosis in panic disorder : an integrative review. Journal of Neuropsychiatry and Clinical Neurosciences. 2001;13:22–34. doi: 10.1176/jnp.13.1.22. [DOI] [PubMed] [Google Scholar]
- Makoroff KL, Cecil KM, Care M, Ball WS., Jr Elevated lactate as an early marker of brain injury in inflicted traumatic brain injury. Pediatric Radiology. 2005;35:668–676. doi: 10.1007/s00247-005-1441-7. [DOI] [PubMed] [Google Scholar]
- Mathews VP, Barker PB, Blackband SJ, Chatham JC, Bryan RN. Cerebral metabolites in patients with acute and subacute strokes: concentrations determined by quantitative proton MR spectroscopy. AJR. American Journal of Roentgenology. 1995;165:633–638. doi: 10.2214/ajr.165.3.7645484. [DOI] [PubMed] [Google Scholar]
- Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR in Biomedicine. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otazo R, Mueller B, Ugurbil K, Wald L, Posse S. Signal-to-noise ratio and spectral linewidth improvements between 1.5 and 7 Tesla in proton echo-planar spectroscopic imaging. Magnetic Resonance in Medicine. 2006;56:1200–1210. doi: 10.1002/mrm.21067. [DOI] [PubMed] [Google Scholar]
- Posse S, Dager SR, Richards TL, Yuan C, Ogg R, Artru AA, Muller-Gartner HW, Hayes C. In vivo measurement of regional brain metabolic response to hyperventilation using magnetic resonance: proton echo planar spectroscopic imaging (PEPSI) Magnetic Resonance in Medicine. 1997;37:858–865. doi: 10.1002/mrm.1910370609. [DOI] [PubMed] [Google Scholar]
- Posse S, Otazo R, Caprihan A, Bustillo J, Chen H, Henry PG, Marjanska M, Gasparovic C, Zuo C, Magnotta V, Mueller B, Mullins P, Renshaw P, Ugurbil K, Lim KO, Alger JR. Proton echo-planar spectroscopic imaging of J-coupled resonances in human brain at 3 and 4 Tesla. Magnetic Resonance in Medicine. 2007;58:236–244. doi: 10.1002/mrm.21287. [DOI] [PubMed] [Google Scholar]
- Prichard J, Rothman D, Novotny E, Petroff O, Kuwabara T, Avison M, Howseman A, Hanstock C, Shulman R. Lactate rise detected by 1H NMR in human visual cortex during physiologic stimulation; Proceedings of the National Academy of Sciences of the United States of America; 1991. pp. 5829–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magnetic Resonance in Medicine. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- Sappey-Marinier D, Calabrese G, Fein G, Hugg JW, Biggins C, Weiner MW. Effect of photic stimulation on human visual cortex lactate and phosphates using 1H and 31P magnetic resonance spectroscopy. Journal of Cerebral Blood Flow and Metabolism. 1992;12:584–592. doi: 10.1038/jcbfm.1992.82. [DOI] [PubMed] [Google Scholar]
- Schurr A. Lactate: the ultimate cerebral oxidative energy substrate? Journal of Cerebral Blood Flow and Metabolism. 2006;26:142–152. doi: 10.1038/sj.jcbfm.9600174. [DOI] [PubMed] [Google Scholar]
- Sijens PE, Levendag PC, Vecht CJ, van Dijk P, Oudkerk M. 1H MR spectroscopy detection of lipids and lactate in metastatic brain tumors. NMR in Biomedicine. 1996;9:65–71. doi: 10.1002/(SICI)1099-1492(199604)9:2<65::AID-NBM397>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Sijens PE, Smit GP, Rodiger LA, van Spronsen FJ, Oudkerk M, Rodenburg RJ, Lunsing RJ. MR spectroscopy of the brain in Leigh syndrome. Brain and Development. 2008;30:579–583. doi: 10.1016/j.braindev.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Tedeschi G, Bertolino A, Campbell G, Barnett AS, Duyn JH, Jacob PK, Moonen CT, Alger JR, Di Chiro G. Reproducibility of proton MR spectroscopic imaging findings. AJNR. American Journal of Neuroradiology. 1996;17:1871–1879. [PMC free article] [PubMed] [Google Scholar]
- Tedeschi G, Bertolino A, Righini A, Campbell G, Raman R, Duyn JH, Moonen CT, Alger JR, Di Chiro G. Brain regional distribution pattern of metabolite signal intensities in young adults by proton magnetic resonance spectroscopic imaging. Neurology. 1995;45:1384–1391. doi: 10.1212/wnl.45.7.1384. [DOI] [PubMed] [Google Scholar]
- Tkac I, Ugurbil K, Gruetter R. On the quantification of low concentration metabolites by 1H NMR spectroscopy in the human brain at 7 tesla; Proceedings of the 10th Meeting of the International Society of Magnetic Resonance in Medicine; Honolulu, USA. 2002. p. 528. [Google Scholar]
- van Rijen PC, Luyten PR, van der Sprenkel JW, Kraaier V, van Huffelen AC, Tulleken CA, den Hollander JA. 1H and 31P NMR measurement of cerebral lactate, high-energy phosphate levels, and pH in humans during voluntary hyperventilation: associated EEG, capnographic, and Doppler findings. Magnetic Resonance in Medicine. 1989;10:182–193. doi: 10.1002/mrm.1910100204. [DOI] [PubMed] [Google Scholar]