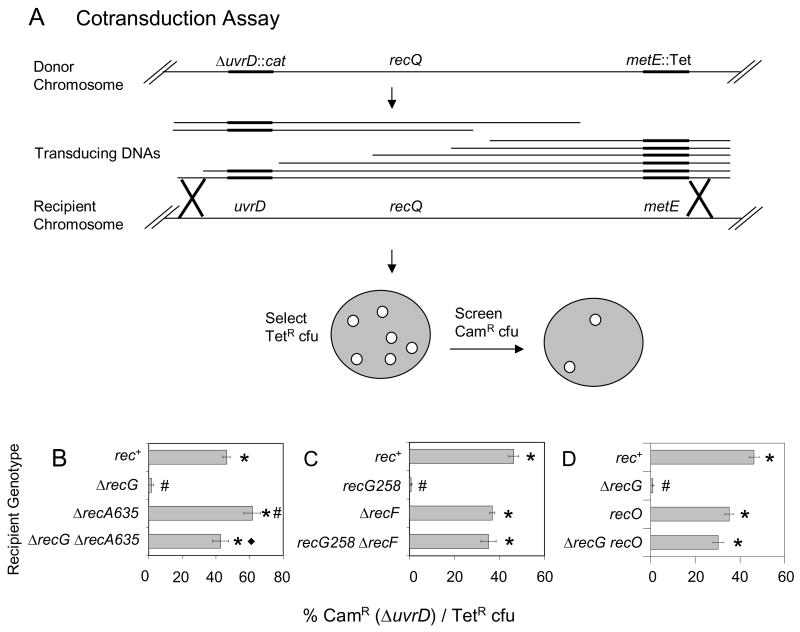
Fig. 1.
Demonstration of inviability of recG ΔuvrD double mutants by cotransduction and requirements for RecA, RecF and RecO for the inviability. (A) Cotransduction assay. Phage P1 grown on a donor strain with two linked antibiotic resistances (SMR9811) recombine into a recipient strain. Transductants are selected for the non-lethal marker (TetR) and screened for those that also contain the potentially lethal marker (ΔuvrD::cat). recQ is located between uvrD and the linked Tet marker, so for examining a requirement for RecQ (Fig. 2) P1 grown on SMR9812, a strain that is ΔrecQ, is used. (B) ΔrecG ΔuvrD cells are inviable and RecA is required for their inviability. ΔuvrD cannot be cotransduced into a ΔrecG recipient (SMR10698), but can into rec+ (SMR10424), ΔrecA (SMR10427) and ΔrecA ΔrecG (SMR10428) recipients carrying pML104-3. (C) RecF is required for recG ΔuvrD inviability. ΔuvrD cannot be cotransduced into recG258 (SMR10419), but can into rec+ (SMR6319), ΔrecF (SMR10423) and ΔrecF recG258 (SMR10437) recipients. (D) RecO is required for ΔrecG ΔuvrD inviability. ΔuvrD cannot be cotransduced into ΔrecG (SMR11133), but can into recO (SMR11134) and recO ΔrecG (SMR11135) recipients. Mean ± SEM of 3 experiments for B, C and D. * indicates a significant difference from recG (B–D). # indicates a significant difference from rec+ (B–D); ◆ indicates a significant difference from the ΔrecA single mutant (B, p = 0.02) of the double mutant indicated.