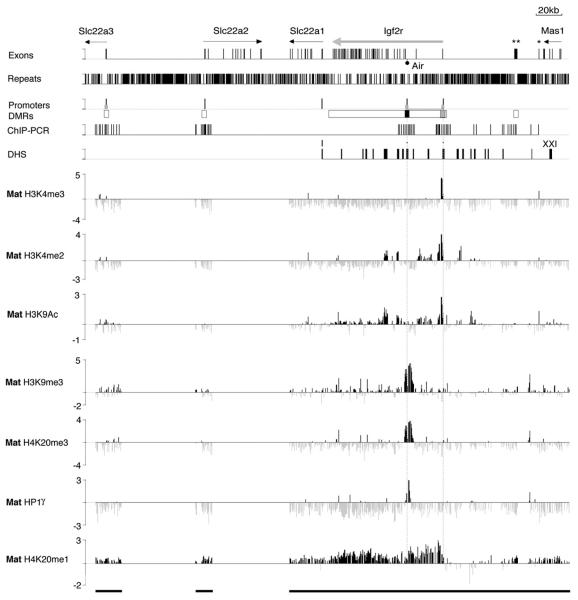
Figure 3. Focal Heterochromatin on the Silent Air Promoter.
ChIP-Chip profile of active (H3K4me3, H3K4me2, H3K9Ac) and repressive (H3K9me3, H4K20me3, and H4K20me1) histone modifications and the heterochromatin protein HP1γ on the maternal (Mat) Igf2r imprinted cluster in MEFB1 cells on the custom PCR tiling array shown in Figure 1B. X axis, 380 kb containing the Igf2r imprinted gene cluster that lacked two regions of 58 kb and 61 kb (gaps in the black bars underneath). Exon track, see Figure 1B. Repeats track, bars show total interspersed repeats (simple repeats, MIR, SINE, LINE1, LINE2, LTRs). Promoters track, bars represent transcription start sites (black) and CpG islands (gray). DMRs track, black and gray boxes; respectively, maternal- and paternal-specific DNA methylated regions; white boxes, regions showing no parental differences in DNA methylation (Stoger et al., 1993). ChIP-PCR track, bars represent 61 sites analyzed by PCR around the Slc22a3, Slc22a2, Igf2r, and Air promoters and Au76-psg. DHS track, bars indicate 21 mapped DHS sites (Pauler et al., 2005). In the seven tracks underneath, each PCR product on the custom tiling array is represented as a single column that is the log2 signal ratio (ChIP/ Input) of the average ratio of medians of eight replicate spots. Vertical dotted lines passing through all tracks mark Air and Igf2r transcription start sites. Only peaks reproducible in biological replicas (Figure S4) were considered positive.