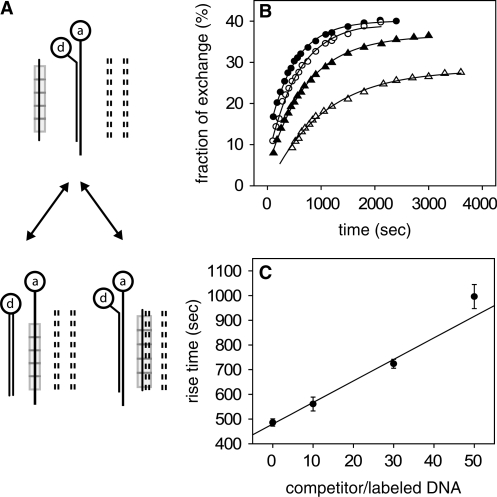
Figure 3.
Effects of double-stranded heterologous competitor on the kinetics of strand exchange between fully homologous ssDNA and dsDNA. (A) Experimental scheme to study RecA-induced recombination in the presence of non-tagged competing DNA duplexes (dashed lines) by FRET: an ssDNA substrate covered with RecA (rectangles) is combined with dsDNA tagged at both strands on the same side of the duplex with a donor-acceptor pair, d and a, respectively. The products include both duplexes having undergone strand exchange as well as synaptic complexes with competitor dsDNA. After strand exchange, no energy transfer takes place. RecA monomers dissociate and are then able to reploymerize on ssDNA. (B) Fraction of tagged duplexes having undergone strand exchange as a function of time for different ratios of competitor to tagged dsDNA concentrations, as obtained from FRET experiments: no competitor (full circles); 10-fold (empty circles); 30-fold (full triangles) and 50-fold (empty triangles). The solid lines are exponential fits to the data. (C) Rise time of the curves in B as function of competitor concentration. The straight line is a linear fit to the data. Error bars represent the uncertainties derived from the fits in B.