Abstract
In DNA site-specific recombination catalysed by tyrosine recombinases, two pairs of DNA strands are sequentially exchanged between separate duplexes and the mechanisms that confer directionality to this theoretically reversible reaction remain unclear. The tyrosine recombinase TnpI acts at the internal resolution site (IRS) of the transposon Tn4430 to resolve intermolecular transposition products. Recombination is catalysed at the IRS core sites (IR1–IR2) and is regulated by adjacent TnpI-binding motifs (DR1 and DR2). These are dispensable accessory sequences that confer resolution selectivity to the reaction by stimulating synapsis between directly repeated IRSs. Here, we show that formation of the DR1–DR2-containing synapse imposes a specific order of activation of the TnpI catalytic subunits in the complex so that the IR1-bound subunits catalyse the first strand exchange and the IR2-bound subunits the second strand exchange. This ordered pathway was demonstrated for a complete recombination reaction using a TnpI catalytic mutant (TnpI-H234L) partially defective in DNA rejoining. The presence of the DR1- and DR2-bound TnpI subunits was also found to stabilize transient recombination intermediates, further displacing the reaction equilibrium towards product formation. Implication of TnpI/IRS accessory elements in the initial architecture of the synapse and subsequent conformational changes taking place during strand exchange is discussed.
INTRODUCTION
Many cellular transactions are mediated by macromolecular machineries in which separate components must be appropriately juxtaposed to bring about a series of ordered molecular events. Progression of the reaction is often coupled to energetically favourable conformational changes, conferring directionality to the process. Well-documented examples of this type of reactions are DNA rearrangements mediated by a variety of specialized recombination systems in prokaryotes and eukaryotes. These rearrangements are involved in a range of biological processes such as gene expression reprogramming, chromosome segregation, the movement of different classes of mobile genetic elements and the life cycle of certain viruses (1).
DNA recombinases, the enzymes that mediate these rearrangements belong to different families of polynucleotidyl transferases that catalyse concerted DNA breakage and rejoining reactions between distant DNA sites (1–3). Members of one large family of these enzymes, the tyrosine recombinase family, cut and reseal DNA through the formation of a transient 3′ phosphotyrosyl DNA-protein bond (4–6). The same reaction mechanism is used by eukaryotic type IB topoisomerases (7,8) and by a class of proteins termed telomere resolvases (or protelomerases) that catalyse the formation of covalently closed DNA hairpins at the ends of bacterial and viral linear replicons (9,10). All these enzymes share a common organization of their catalytic domain, with a similar constellation of conserved active-site residues, including the tyrosine that provides the primary nucleophile for DNA cleavage.
However, the complexity of the reaction catalysed by these different proteins depends on their biological function. Type IB topoisomerases function as a monomer to release excess topological strain in DNA, whereas hairpin formation by telomere resolvases requires the collaboration of two molecules to cleave and rejoin both DNA strands in a duplex (9,10). Tyrosine recombinases perform the most intricate reaction of all, which is the reciprocal exchange of two pairs of DNA strands between separate recombination sites (4–6). Coordination of the DNA cleavage and rejoining reactions takes place within a synaptic complex containing four recombinase molecules assembled onto short (∼30 bp) DNA segments, termed the ‘core’ recombination sites (Figure 1). The core site is usually composed of inversely oriented recombinase binding elements surrounding a 6- to 8-bp spacer sequence (or ‘overlap’ region) at the borders of which DNA cleavages take place. In this complex, one specific pair of recombinase molecules catalyses the first strand exchange, generating a Holliday junction (HJ) intermediate that is then resolved to recombinant products by the other pair of subunits (Figure 1). Each strand exchange is a two-step transesterification reaction in which the 3′ phosphotyrosyl DNA–protein bond produced by cleavage of one recombination site is attacked by the 5′ OH end released in the opposite site. DNA strands are exchanged by melting 3–4 bp from the spacer sequence adjacent to the cleavage sites and re-annealing them to the partner complementary strand in order to adequately orient the 5′ OH ends for ligation (11). This ‘homology-testing’ step of recombination ensures that appropriate strands are exchanged during the reaction, thereby preventing formation of aberrant products (Figure 1).
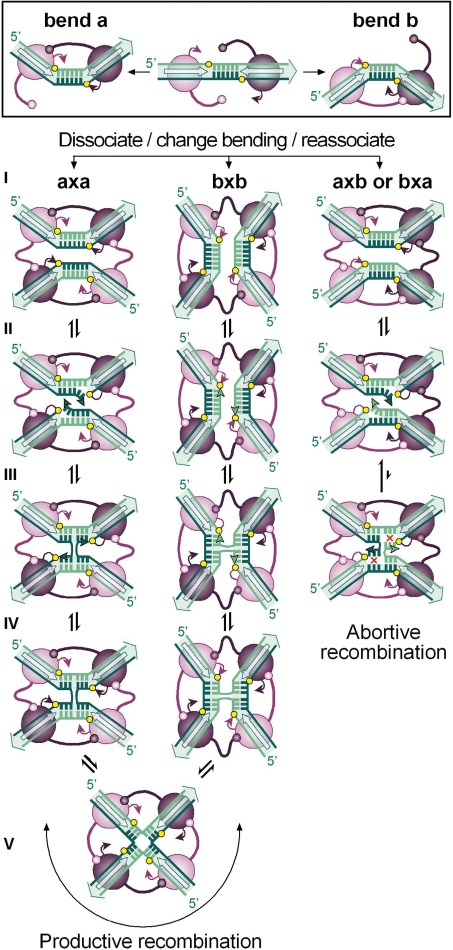
Figure 1.
The strand-exchange reaction catalysed by tyrosine recombinases. Binding of a pair of recombinase molecules (purple ovals) onto the core recombination site recognition elements (open arrows) may bend DNA to opposite directions (a or b) exposing one or the other strand for catalysis (coloured dark and light green, respectively). In all cases studied, activity of the recombinase subunits is controlled by allosteric interactions in which the active subunit donates its C-terminus (represented by a stem-and-ball extension) to its neighbour. During synaptic complex formation (I), both recombination sites may pair with an antiparallel alignment if they are in the same bent configuration (a × a or b × b) or with a parallel alignment if they are bent in opposite directions (a × b or b × a). In all cases, this leads to the formation of a recombinase tetramer in which diagonally opposed subunits are activated, while the other pair of subunits is inactive. Antiparallel pairing of the recombination sites proceeds to strand exchange through a sequence of reversible steps during which the exposed strands are cleaved (II), exchanged (III) and then rejoined (IV) to the partner strand to form a HJ. Isomerization of the HJ (V) inactivates one pair of recombinase subunits and activates the other pair of subunits to exchange the second pair of strands. For each strand-exchange reaction, the recombinase catalytic tyrosine (purple arrowhead) attacks the adjacent sessile phosphate (yellow circle) to form a phosphotyrosyl bond that is in turn attacked by the 5′OH end of the cleaved DNA strand (green arrowhead). Note that recombination that initiates with one configuration of the synapse terminates with the core sites in the opposite configuration and reciprocally. Parallel pairing of the recombination sites generally leads to abortive recombination due to the lack base pairs complementarity (red crosses) in the overlap region (step III).
This mechanism of sequential strand exchange implies that separate pairs of recombinase molecules are active at both steps of the reaction. For the different recombinases that have been studied in detail, isomerisation of the complex is proposed to be controlled by allosteric interactions between adjacent recombinase subunits in the tetramer so that diagonally opposed active sites are switched on and off during recombination (12–16). However, every step in the tyrosine recombinase pathway is theoretically reversible and it is currently difficult to predict which DNA strands will be cleaved first and what factors may influence individual equilibriums of the strand-exchange reaction to ensure efficient substrate conversion to product (Figure 1; 17).
The tyrosine recombinase TnpI encoded by the transposon Tn4430 from Bacillus thuringiensis mediates recombination at the internal resolution site (IRS) of this element to resolve cointegrate molecules arising from replicative transposition (18,19). In other transposons of the same family (i.e. the Tn3 family), this function is carried out by a resolvase protein belonging to a different family of enzymes, the serine recombinase family (5). The IRS of Tn4430 (116 bp) contains four TnpI-binding motifs dividing the recombination site into functionally separate regions (Figure 2A). The IR1 and IR2 inverted motifs form the core recombination site where TnpI mediates strand exchange, whereas the adjacent tandem motifs DR1 and DR2 are dispensable accessory elements to which additional regulatory TnpI subunits bind, thereby forming part of the active recombination complex. Formation of this complex stimulates recombination of DNA molecules that carry directly repeated IRSs (19). On circular DNA substrates, this reaction exclusively produces two-noded catenane molecules in which both product rings are singly interlinked (Figure 2B). This implies that the DR1 and DR2 accessory motifs are involved in the formation of a topologically defined synaptic complex in which the two recombination sites are interwrapped in a specific manner. This complex only forms readily between sites in a head to tail configuration, thereby acting as a ‘topological filter’ to ensure resolution selectivity of the reaction. This mechanism of selectivity is important for the biological function of the TnpI/IRS recombination system, which is to resolve transposition cointegrates at the time they form without causing any deleterious DNA rearrangements. In the absence of DR1 and DR2, TnpI-mediated recombination at the minimal IR1–IR2 core site displays ‘relaxed’ activity, giving rise to topologically variable products following random collision of the recombination sites (Figure 2B; 19).
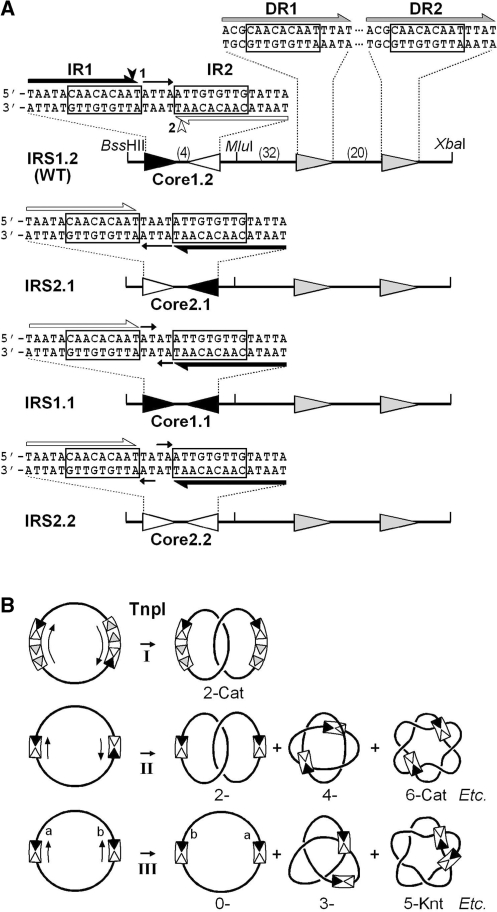
Figure 2.
The TnpI/IRS resolution system of Tn4430. (A) Structure of the IRS recombination site and its derivatives. Wild-type IRS (IRS1.2, 116 bp) contains two 14-bp inverted motifs, IR1 (filled triangle) and IR2 (open triangle), forming the recombination core site; and two 16 bp accessory TnpI-binding motifs, DR1 and DR2 (shaded triangles), playing a regulatory role in recombination. All motifs share a common sequence of 9 bp (boxed) thought to be bound by separate recombinase molecules within the recombination complex. The distance that separates the different motifs (in bp) is indicated. TnpI-mediated DNA cleavages at IR1 and IR2 take place at symmetrical positions staggered by 6 bp as indicated by a filled and open arrowhead, respectively. Both motifs are separated by a spacer sequence of 4 bp (orientation shown by an arrow). The IRS2.1 variant was constructed by reversing the orientation of the IR1–IR2 core site (Core1.2) with respected to the DR1–DR2 accessory motifs. IRS1.1 and IRS2.2 were constructed by replacing the IR1–IR2 core site with symmetrical sequences containing inversely oriented IR1 (Core1.1) and IR2 (Core2.2) half sites, respectively. Simple core recombination sites corresponding to the different IRS variants were made by replacing the DR1–DR2 sequence with an unrelated DNA fragment of the same length (see the ‘Materials and Methods’ section). (B) Topologically constrained and relaxed recombination activity of TnpI. TnpI-mediated recombination of supercoiled DNA molecules carrying directly repeated full-length IRSs exclusively generates two-noded catenane products (2-Cat) following the formation of a topologically defined synaptic complex involving the accessory motifs DR1 and DR2 (I). In the absence of DR1-DR1, recombination between minimal IR1–IR2 core sites takes place after random collision, trapping a variable number of DNA supercoils between both recombination sites and generating a mixture of catenanes (x-Cat) containing an even number of crossings (or nodes) if the two sites are in direct repeat (II), or a mixture of unknotted and knotted inversion products (x-Knt) containing an odd number of nodes if the two sites are in inverted repeat (III).
Here, we show that the accessory DR1–DR2 motifs and their bound TnpI protomers not only provide an architectural scaffold for the formation of the topologically constrained TnpI/IRS recombination complex, They also directly contribute to the directionality and efficiency of strand exchange (i) by imposing a specific order of activation of the catalytic TnpI subunits of the complex and (ii) by stabilising subsequent intermediates of the reaction. Asymmetry in the IR1–IR2 core site sequence plays a secondary role in this process by reinforcing the directionality imposed by the accessory sequences and proteins.
MATERIALS AND METHODS
DNA substrates and proteins
Plasmid pGIC002 carries a wild-type IRS derivative (IRS1.2) containing an MluI site between the IR1–IR2 core site and the DR1–DR2 accessory sequences (Figure 2A; 19). In pGIV023, a DNA fragment of unrelated sequence was substituted for the pGIC002 fragment carrying DR1 and DR2 (19). The recombination site variants containing the reversed or symmetrical cores joined to the DR1–DR2 accessory motifs, or not, were constructed by swapping the Core1.2 sequence of pGIC002 and pGIV023 with 46 bp BssHII-MluI linkers containing the modified Core2.1, Core1.1 and Core2.2 sequences (Figure 2A). The resulting plasmids were used to construct reporter plasmids containing pairs of recombination sites separated by a 1.3-kb kanamycin resistance cassette as previously described (19).
Plasmid pGIV004 is a pCB104 derivative encoding the TnpI protein under Escherichia coli Plac promoter. Plasmid pGIV007 is a pBAD/HisA-based vector (Invitrogen) expressing an N-terminal fusion between TnpI and a (His)6 tag (19). The TnpI-H234L mutant was constructed by unique restriction site elimination mutagenesis (20) using pGIV007 as a template and the oligonucleotides 5′-TTATTACTCCACTCCAATTACGACA (changing the His234 codon CAC into Leu codon CTC) and 5′-AGTTAAGCCAGTTAACACTCCGCTAT (removing the pGIV007 AccI site) as primers. The N-terminally (His)6-tagged TnpI-H234L protein encoded by the resulting pCNL08 vector was purified as previously described for wild-type TnpI (19).
In vivo recombination
TOP10 E. coli cells (Invitrogen) harbouring the reporter plasmids were transformed with the TnpI expression vector pGIV004 or with pCB104. Plasmid DNA was then extracted from a pool of colonies and analysed by electrophoresis on a 0.7% agarose gel in TAE buffer (20).
In vitro recombination
Recombination on supercoiled plasmids and DNase I nicking reactions were carried out as previously described (19). Reactions were run on 0.8% agarose TAE electrophoresis gels and analysed using Quantity OneTM software (Bio-Rad) after SYBR green I staining (SIGMA) and UV illumination.
Characterization of Holliday junction intermediates
In vitro, recombination reactions were carried in the presence of the hexapeptide KWWCRW (21) at a final concentration of 2 µM. After cleavage with AvaII, HJ-containing χ-forms were purified from a 1% agarose gel and 5′-end-labelled with γ32P-ATP (Amersham). Labelled χ-forms were left uncut or recleaved with NruI and ScaI and analysed by electrophoresis on 1% agarose, 50-mM NaOH, 1-mM EDTA denaturing gels. Gels were dried and exposed to X-ray films for autoradiography.
Mapping of sequential DNA cleavages by primer extension
Following in vitro recombination of pGIC002 with TnpI-H234L, unreacted substrate and cleaved products were separated on a 0.8% agarose gel and purified using Genelute agarose spin columns (SIGMA). Primer extension analysis was performed with 5′ 32P-labelled primer 1 (5′-TCAGCAACACCTTCTTCACGAGG, hybridising 193 nt upstream of IR1 cleavage site) and primer 2 (5′-ATTAATGTGAGTTAGCTCACTC, hybridising 254 nt upstream of IR2 cleavage site) using thermosequenase (USB). Extension products were analysed on 6% polyacrylamide sequencing gels (20) alongside DNA sequencing reactions performed with the same primers. Gels were dried and exposed to X-ray films.
RESULTS
Directly repeated DR1–DR2 accessory sequences can impose resolution selectivity to inversely oriented IR1–IR2 core sites in vivo
Asymmetry within Tn4430 IRS is primarily conferred by the presence of the accessory motifs DR1 and DR2, and by the 4-bp spacer sequence (5′-ATTA-3′) that separates the perfectly symmetrical core site motifs IR1 and IR2 (Figure 2A). To see whether the relative orientation of both asymmetrical elements could influence recombination, an IRS variant (IRS2.1) was constructed by reversing the orientation of the IR1–IR2 core site with respect to the DR1–DR2 accessory sequences (Figure 2A). This new site was used to construct two recombination reporter plasmids (Figure 2). One plasmid (pIRS2.1 × 2.1) carries tandem copies of IRS2.1, while the other plasmid (pIRS1.2 × 2.1) contains one copy of the wild-type IRS (IRS1.2) and one copy of IRS2.1 (Figure 3). In the latter substrate, the DR1–DR2 accessory sequences of both recombination sites are in the same orientation, while the IR1–IR2 core sites are reversed with respect to each other.
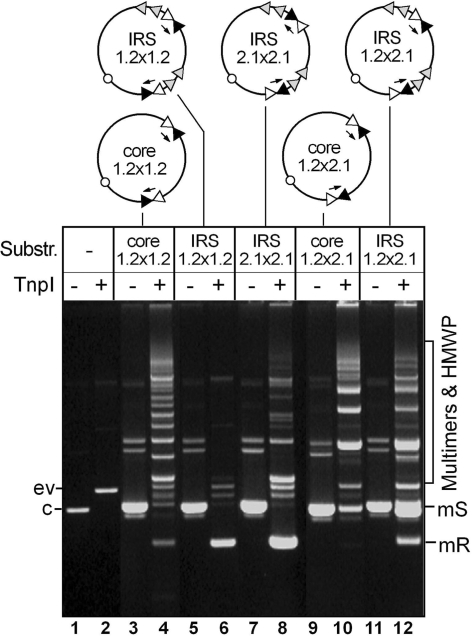
Figure 3.
TnpI-mediated relaxed and constrained recombination between directly and inversely repeated core sites in vivo. Reporter plasmids containing different arrangements of the IR1–IR2 core sites joined to directly repeated DR1–DR2 sequences or not were transformed into E. coli TOP10 cells harbouring the TnpI expression vector pGIV004 (ev) or the control plasmid pCB104 (c). Plasmid DNA was isolated from a pool of transformants and run uncut on a 0.8% agarose gel. Intramolecular recombination of the substrate monomer (S) yielded monomeric resolution (mR) products in which the DNA fragment between both recombination sites was deleted. Unconstrained recombination generated multimers of the substrate and/or resolution product, together with a mixture of high molecular weight products arising from both inter and intramolecular recombination reactions. Reporter plasmids are diagrammed on the top of the figure, with the IR1 and IR2 core motifs represented by filled and open triangles, respectively; and the DR1 and DR2 accessory motifs by shaded triangles. Arrows show the orientation of the core sites as determined by the central spacer sequence.
Recombination of these plasmids was first examined in vivo, in E. coli cells expressing the TnpI recombinase from a separate vector. The results were compared to those obtained for equivalent reporter plasmids carrying directly repeated copies of the wild-type IRS (pIRS1.2 × 1.2) or two copies of the minimal IR1–IR2 core site, either in direct or inverted repeat (pCore1.2 × 1.2 and pCore1.2 × 2.1, respectively; Figure 3). As previously reported (19), TnpI-mediated recombination of pCore1.2 × 1.2 was non-selective, generating a ladder of products arising from both intra and intermolecular recombination (Figure 3, lane 4). In contrast, the wild-type IRS substrate pIRS1.2 × 1.2 was resolved to completion by intramolecular recombination owing to the presence of the DR1–DR2 accessory sequences (lane 6). Complete deletion of the DNA fragment between both recombination sites was also observed for the pIRS2.1 × 2.1 reporter plasmid carrying directly repeated copies of the reversed core site variant IRS2.1 (lane 8). However, in this case, multimers of the deletion product were still present in the reaction, indicating that resolution selectivity conferred by the accessory sequences was only partial, unable to completely reduce multimeric forms that were produced by intermolecular recombination (Figure 3, compare lanes 6 and 8).
Plasmid multimers arising from ‘relaxed’ recombination were also generated with the inversion substrate pCore1.2 × 2.1 and the pIRS1.2 × 2.1 plasmid carrying one copy of wild-type IRS and one copy of the reversed core site variant IRS2.1 (Figure 3, lanes 10 and 12). Unexpectedly, recombination of pIRS1.2 × 2.1 also produced substantial amounts of intramolecular deletion products that were not seen with pCore1.2 × 2.1. This shows that the presence of directly repeated DR1–DR2 sequences can impose resolution selectivity to inversely oriented core sites in vivo, poising the IR1-bound TnpI subunits for exchanging DNA strands with the IR2-bound subunits. To perform this reaction, the two sites must be brought in a parallel alignment and strand exchange would then generate mismatched base pairs in the overlap region (see Figure 1). Formation of this kind of products is expected to be strongly disfavoured unless some mechanism would stabilize the strand-exchange intermediate promoting its processing into recombination products.
Recombination of the different substrates was also examined in vitro. Supercoiled plasmids carrying pairs of recombination sites were incubated with purified TnpI and reactions were analysed by agarose gel electrophoresis after digestion with AvaII (Figure 4). The reporter substrates carrying directly repeated IRS1.2 or IRS2.1 sites recombined to give specific deletion products (D1 and D2) as expected for intramolecular recombination between both sites. Recombination of both substrates also generated a slower migrating DNA species corresponding to the HJ intermediate of the reaction. However, recombination at the reversed core site variant IRS2.1 occurred at a much lower rate than recombination at wild-type IRS1.2 (Figure 4). This result is consistent with the lower activity of IRS2.1 observed in vivo (Figure 3), and shows that changing the orientation of the core-site central region affects the efficiency of the strand-exchange reaction catalysed by TnpI.
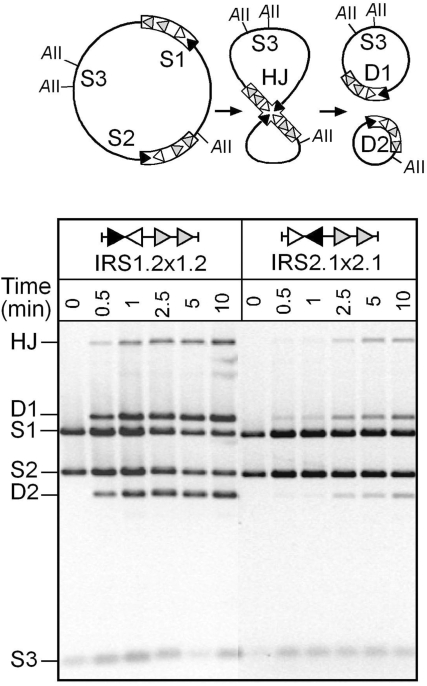
Figure 4.
TnpI-mediated recombination at the reversed core site IRS2.1 variant in vitro. Supercoiled plasmids carrying directly repeated copies of wild-type IRS (IRS1.2 × 1.2) or modified IRS in which the orientation of the IR1–IR2 core site is reversed with respect to the DR1–DR2 accessory sequence (IR2.1 × 2.1) were incubated for the indicated times with purified TnpI. Reaction were digested with AvaII (AII) and analysed by electrophoresis on a 0.8% agarose gel. S1, S2 and S3 are the AvaII fragments from the initial substrates. D1 and D2 are specific fragments from the deletion products. AvaII digestion of the reactions also produced χ-forms corresponding to the Holliday junction (HJ) intermediate of recombination.
Neither inversion nor deletion products were observed when the inversely oriented core-site substrate pIRS1.2 × 2.1 was incubated with TnpI under standard conditions (data not shown). However, as detailed below, detectable levels of HJ products accumulated in the presence of inhibitor peptides known to block specific steps of the tyrosine recombinases pathway. This supports the idea that the presence of the accessory sequences provides a topological filter mechanism that prevents random synapsis as observed with simple recombination core-site substrates (19), and that formation of the accessory motifs-containing complex may lead to strand-exchange products between misaligned core sites that do not form, or are highly unstable, under normal conditions. In vivo, these intermediates may be resolved to fully recombined products by host-dependent DNA repair mechanisms as was proposed for other recombination systems (16,22–24).
The DR1–DR2 accessory sequences impose a specific alignment of the recombination core sites within the synaptic complex
To further investigate the respective contribution of the IRS core and accessory sequences to the alignment of the recombination sites, perfectly symmetrical core sites were made by changing the IR1–IR2 spacer sequence 5′-ATTA-3′ to 5′-ATAT-3′ and 5′-TATA-3′. The resulting Core1.1 and Core2.2 variants consist of inversely oriented IR1 and IR2 half sites, respectively (Figure 2A).
In the absence of DR1–DR2, in vitro recombination between directly repeated Core1.2 sites only yielded deletion products (Figure 5, panel I). In contrast, recombination of substrates containing pairs of symmetrical Core1.1 and Core2.2 sites generated specific bands arising from both deletion (D1 and D2) and inversion (Inv1 and Inv2) reactions (Figure 5, panels II and III). Additional product bands resulting from left-to-left and right-to-right joining of the recombination sites flanking sequences were also detected. These products (labelled ‘Inter’ in Figure 5) are intermolecular recombination products that appeared at a later stage of recombination. They were found to accumulate at the expense of relaxed DNA molecules produced by type-II-topoisomerase activity inherent to the inversion reaction (data not shown) (25,26). The reason why intermolecular recombination products only form efficiently on relaxed DNA substrates remains unclear and is not within the scope of this study. Whichever this reason, the recombination pattern obtained for the pCore1.1 × 1.1 and pCore2.2 × 2.2 substrates is fully consistent with recombination occurring with all possible alignments of the recombination sites due to the lack of asymmetry in the overlap region. This pattern sharply contrasts with that given by the pIRS1.1 × 1.1 and pIRS2.2 × 2.2 plasmids, in which the symmetrical Core1.1 and Core2.2 sites are under the control of directly repeated DR1–DR2 accessory sequences (Figure 5, panels V and VI). As for the wild-type pIRS1.2 × 1.2 substrate (panel IV), recombination of both plasmids only generated deletion products. This shows that the presence of DR1 and DR2 specifies all the required information to impose resolution selectivity to recombination by forcing a specific alignment of the core sites during synaptic complex assembly.
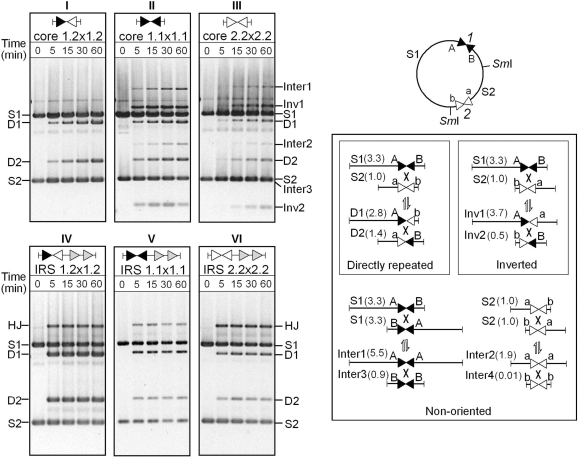
Figure 5.
TnpI-mediated recombination at symmetrical core sites in vitro. Reporter plasmids carrying tandem copies of the wild-type (Core1.2) or symmetrical (Core1.1 and Core2.2) core sites under the control of DR1–DR2 accessory sequences (panels IV–VI) or not (panels I–III) were incubated with purified TnpI for the indicated times. Reactions were digested with SmaI (SI) and analysed by agarose gel electrophoresis. The cartoon summarizes all possible DNA fragments that may arise from recombination at both substrate sites (site1 and 2) after digestion with SmaI. S1 and S2 are the initial substrate fragments. D1 and D2 are DNA fragments arising from recombination between directly oriented sites1 and 2. Inv1 and Inv2 are recombination products of inversely oriented sites. Inter1, Inter2, Inter3 and Inter4 are specific ‘intermolecular’ site1 × site1 and site2 × site2 recombination products in which flanking DNA segments are joined in head-to-head and tail-to-tail configuration (Inter4 was too small to be seen on the gel). HJ intermediates migrate as a χ-form at an upper position of the gel.
In addition to altering recombination outcome, changes made in the central region appeared to affect the efficiency of strand exchange. In the absence of DR1–DR2, recombination at the symmetrical Core2.2 site was much less efficient than recombination at the reciprocal Core1.1 site (Figure 5, compare panels II and III). As seen for the wild-type IRS, addition of the DR1–DR2 accessory sequences was found to increase the initial rate of recombination at both symmetrical core-site variants with concomitant appearance of HJ products that are never detected with the core sites alone (Figure 5). However, HJ intermediates accumulated to a proportionally greater extent with the pIRS2.2 × 2.2 substrate than with the pIRS1.1 × 1.1 or pIRS1.2 × 1.2 substrates (Figure 4, compare panel VI with panels IV and V).
The DR1–DR2 accessory sequences impose a unique topology to the synaptic complex, irrespective of the sequence and orientation of the core sites
Recombination of a circular DNA molecule carrying directly repeated copies of wild-type IRS exclusively produces two-noded catenane products as a consequence of the specific arrangement of the recombination sites in the complex (19; Figure 2). In order to determine whether changes made in the core site could influence the geometry of the synapse, the topology of the products arising from recombination at the different IRS variants was examined (Figure 6).
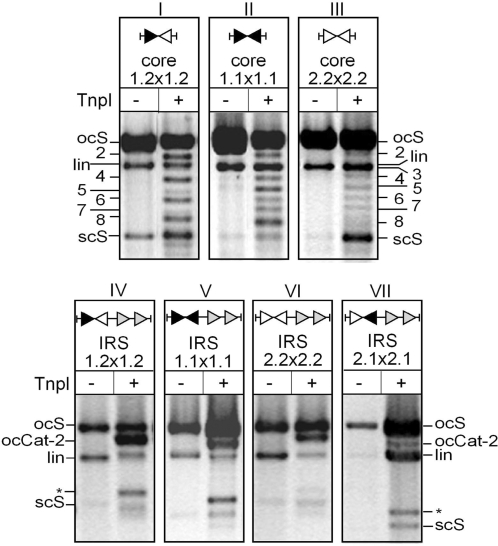
Figure 6.
DNA topology of recombination products. Recombination of circular substrates containing pairs of core site variants in the absence (panels I–III) or presence (panels IV–VII) of directly repeated DR1–DR2 accessory motifs were treated with TnpI or left untreated as indicated. Reactions were singly nicked with DNase I and run on a 0.8% agarose gel. Positions of the open circular (ocS), linear (lin), and supercoiled (scS) forms of the substrates are indicated, as are the positions of the nicked two-, three-, four-, five-, six-, seven- and eight-noded catenane and knot products of recombination. Note that the majority of secondary (twist) knots generated by consecutive rounds of recombination at Core1.2 (panel I) migrate slightly ahead of the primary (torus) knots arising from recombination at the symmetrical Core1.1 and Core2.2 sites (panels II and III). Recombination of the DR1 and DR2-containg plasmids (panels IV–VII) yielded two-noded catenanes (ocCat-2) as the only product of recombination. Additional bands (asterisk) correspond to circular deletion products that were released from catenanes by over digestion with DNase I.
As was previously shown, unconstrained recombination at the IR1–IR2 core site gave a mixture of even-noded catenane products in which the deletion products are interlinked a variable number of times (19). This is consistent with recombination occurring after random collision of the recombination sites, trapping a variable number of supercoils from the initial substrate during synapsis (Figure 2B). Small amounts of knots were also observed in the reaction. These are secondary products arising from the conversion of the initial catenanes following consecutive rounds of recombination (Figure 6, panel I; 19). In contrast, recombination at the symmetrical Core1.1 and Core2.2 sites generated both catenanes and knots as primary products of the reaction, confirming that recombination has lost orientation selectivity, mediating both deletion and inversion reactions on the initial substrate (Figure 6, panels II and III).
However, when directly repeated copies of DR1–DR2 were joined to the symmetrical Core1.1 and Core2.2 sites, recombination exclusively yielded two-noded catenanes as observed for the wild-type IRS (Figure 6, panels IV–VI). The two-noded catenane was also the only recombination product detected for the pIRS2.1 × 2.1 substrate in which the orientation of the core site is reversed with respect to the accessory sequences (Figure 6, panel VII). This indicates that the DR1–DR2 accessory sequences are major determinants for the formation of a topologically defined synapse in which a fixed number of DNA supercoils are trapped, and that the architecture of the productive synapse remains the same whichever the changes made in the core sites.
The DR1–DR2 accessory sequences impose a specific order of strand exchange to the recombination reaction
The above results support the idea that the accessory TnpI-binding motifs DR1 and DR2 specify resolution selectivity by determining the architecture of the synapse and by imposing a specific alignment of the core sites within the recombination complex. A further question is whether this architecture of the synaptic complex determines a specific order in the strand-exchange reaction catalysed by TnpI. To answer this question, we determined which DNA strands were exchanged in the HJ intermediates obtained for each version of the recombination site (Figure 7).
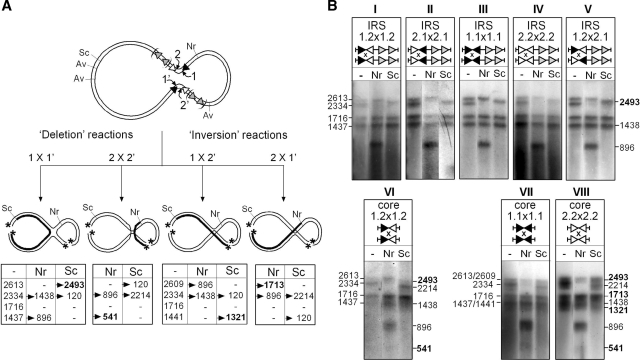
Figure 7.
Analysis of HJ intermediates formed at different variants of IRS. HJ products that accumulated in recombination reactions performed with different IRS substrates (see the ‘Materials and Methods’ section) were cut with AvaII (Av), gel purified, and 5′-labelled with 32P (asterisk). The resulting χ-forms were left untreated (−) or recut with NruI (Nr) or ScaI (Sc) and run on DNA denaturing gels. (A) Different patterns of radio-labelled single-stranded DNA fragments are expected depending on whether the HJ is formed by the IR1-bound TnpI subunits (1 × 1′), the IR2-bound subunits (2 × 2′) or opposite combinations of IR1- and IR2-bound subunits (1 × 2′ and 2 × 1′, respectively). Sizes (in nt) of the diagnostic fragments obtained for the different possibilities of strand exchange are reported in the corresponding tables. One fragment (in bold) is specific for each possibility and the corresponding DNA strand is shown as a thick line in the cartoons showing the different HJ isoforms. (B) Bands patterns obtained for the different substrates after autoradiography of the gels. For each group of substrates, sizes of the initial χ-form fragments are indicated on the left, and the new fragments that were generated after NruI and ScaI cleavage are shown on the right. All the DR1–DR2-constraining substrates (panels I–V) show a unique pattern of bands corresponding to the 1 × 1′ exchange, irrespective of the nature and orientation of the core sites. The pattern obtained for the pCore1.2 × 1.2 substrate (panel VI) is consistent with strand exchange occurring at both IR1 and IR2. HJ formation at the symmetrical Core1.1 and Core2.2 sites (panel VII and VIII) occurred with all possibilities of strand exchange.
Under standard conditions, in vitro recombination of most substrates only gave weak accumulation of HJ intermediates. In particular, no HJ was detected for the simple core-site substrates (Figure 5) or for the pIRS1.2 × 2.1 reporter plasmid (see above). For all substrates, the yield of HJ intermediates could be substantially increased by performing the reactions in the presence of a six-amino acid peptide (KWWCRW) that was previously identified for blocking specific steps of the recombination reaction catalysed by lambda integrase (λ Int) and several other enzymes of the tyrosine recombinase family (21,27) (data not shown).
Analysis of the HJ formed by recombination between directly repeated wild-type IRSs revealed that one specific pair of DNA strands was exchanged by the recombinase (Figure 7B, panel I). Digestion of the χ-form with NruI and ScaI yielded radio-labelled single-stranded DNA fragments of 1438 and 2493 nt, respectively. These bands are specific for strand exchange occurring at IR1 (Figure 7A, 1 × 1′). The expected products for strand exchange at IR2 (2 × 2′) were not detected. This result indicates that recombination at wild-type IRS proceeds with a defined order, with the IR1-bound TnpI subunits exchanging the first pair strands to form the HJ intermediate.
This ordered process could be dictated, either by an intrinsic feature of the IR1–IR2 core site or by the geometry imposed by the accessory motifs within the recombination complex. Supporting the latter possibility, digestion of the HJ produced by recombination at the reversed core-site variant IRS2.1 showed the same pattern as that seen for IRS1.2, indicating in this case that strand exchange was catalysed by the IR2-bound subunits (Figure 7B, panel II). Likewise, recombination of the pIRS1.1 × 1.1, pIRS2.2 × 22 and pIRS1.2 × 2.1 substrates generated the same 1 × 1′ form of the HJ intermediate, in which strand exchange was catalysed by the TnpI protomers bound to the most distal core-site motif with respect to the accessory sequences (panels III–V). Thus, formation of the synaptic complex containing the accessory motifs DR1 and DR2 imposes a specific positioning of the recombination core sites in the complex such that one specific pair of recombinase subunits (those bound to the distal core-site motifs) are activated first to initiate recombination, irrespective of the sequence and orientation of the core sites. For the pIRS1.2 × 2.1 substrate, this brings the inversely oriented core sites in a normally unproductive parallel alignment generating HJ products with unpaired base pairs in the overlap region (Figure 1).
Further demonstrating that it is the geometry imposed by DR1 and DR2 that determines the specific alignment of the core sites in the synapse, recombination at the minimal IR1–IR2 core site yielded a mixture of HJ products resulting from strand exchange at both IR1 (1 × 1′) and IR2 (2 × 2′) (Figure 7B, panel VI). The pattern of bands obtained for the symmetrical Core1.1 and Core2.2 sites was even more complex, showing strand-exchange products arising from all possible alignments of the recombination sites (1 × 1′, 2 × 2′, 1 × 2′ and 2 × 1′), which is consistent with complete loss of selectivity (Figure 6B, panels VII and VIII).
Decision of the order of strand exchange takes place before cleavage
Although the HJ analysis reported in the above section provides good evidence that TnpI mediates recombination with a specific order of strand exchange, it does not totally rule out the possibility that recombination may occur using alternative pathways, one of which generates more stable HJ intermediates than the other giving rise to a greater accumulation in the reactions. To clarify this, the order of activation of TnpI subunits was determined for a complete recombination reaction by trapping the covalent intermediates that are generated at both cleavage steps (Figure 8).
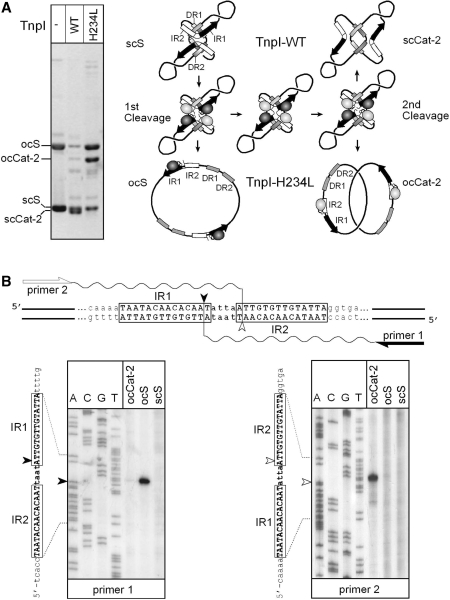
Figure 8.
Ordered DNA cleavages during TnpI-catalysed sequential strand exchanges (A) TnpI and TnpI-H234L recombination pathways. A reporter plasmid carrying directly repeated IRSs was incubated with wild-type TnpI (TnpI-WT) or TnpI-H234L, and the reactions were run uncut on a 0.8% agarose gel. Both proteins assembled onto the supercoiled substrate (scS) to form the TnpI/IRS-specific synapse. For clarity, only one of both possible topological organizations of the synaptic complex is shown (19). The IR1–IR2 core sites and the DR1 and DR2 accessory motifs are represented by black/white arrows and shaded boxes, respectively. The core TnpI subunits bound to IR1 and IR2 half sites are shown as dark and light shaded ovals, respectively. Sequential strand-exchange reactions catalysed by TnpI-WT leads to fully supercoiled two-noded catenane products (scCat-2). In the presence of TnpI-H234L, DNA rejoining was partially inhibited leading to the formation of open circular forms of the substrate (ocS) and catenane product (ocCat-2). (B) Mapping of TnpI-H234L DNA cleavages. Gel-purified products arising from the first (ocS) and second (ocCat-2) DNA cleavage reactions were used as a template in primer extension analysis to determine whether cleavage occurred at IR1 (primer 1) or IR2 (primer 2). Extension products were analysed on a 6% sequencing gel alongside of a DNA sequencing reaction performed with the same primers. Extensions performed with unreacted substrate (scS) were used as a control in the experiment. IR1 and IR2 cleavage positions are shown by a filled and open arrowhead, respectively. Note that the thermostable DNA polymerase used in the experiment added an extra dA nucleotide at the 3′ end of the extension products.
To this end, recombination was performed with a TnpI catalytic mutant, TnpI-H234L, in which the second histidine of the tyrosine recombinases catalytic motif ‘R-K-H-R-H/W-Y’ (7) was changed to leucine. In the yeast recombinase Flp, the equivalent mutation led to a defect in DNA rejoining, giving rise to protein-DNA complexes accumulation (28,29).
Under normal conditions, TnpI-mediated recombination between directly repeated IRSs generates fully supercoiled two-noded catenane products that migrate slightly below the substrate in an agarose gel (Figure 8A). A different pattern was observed with TnpI-H234L. In this case, recombination generated two major products corresponding to the relaxed (open circular) forms of the substrate and of the two-noded catenane, respectively (Figure 8A). This pattern is consistent with TnpI-H234L being partially affected in DNA rejoining. The reaction sometimes stopped after the first strand cleavage, leading to nicked substrate accumulation, whereas other times it proceeded through the second strand exchange, releasing catenane products in which both recombinant circles were relaxed (Figure 8A).
To determine which specific pairs of DNA strands were cleaved at both recombination steps, the nicked forms of the substrate and catenane products were purified from an agarose gel and subjected to primer extension analysis (Figure 8B). When the nicked form of the substrate was used as a template, a strong band corresponding to cleavage at IR1 was observed (Figure 8B, primer 1). No specific extension product was detected in the reaction aiming at mapping cleavage at IR2 (primer 2). Conversely, isolated catenane products showed strong cleavage at IR2, whereas cleavage at IR1 was essentially undetectable (Figure 8B).
These results confirm and extend the conclusion that TnpI-mediated recombination at IRS occurs with a specific order of strand exchange, and that the HJs that were analysed in the above section are actual intermediates of the reaction. They also indicate that the specific order of activation of the TnpI catalytic subunits is determined before the initial cleavage step of recombination since no alternative cleavages of the substrate were detected in the primer extension analysis.
DISCUSSION
The data presented here show that the accessory elements of Tn4430 TnpI/IRS resolution system act at multiple steps to control recombination. They provide an architectural scaffold for the assembly of a topologically and functionally selective synapse, and function as allosteric effectors of DNA-strand-exchange catalysis to orient the reaction.
The TnpI/IRS accessory elements impose a specific geometry to the catalytic recombinase tetramer in the synaptic complex
Recombination performed with the symmetrical Core1.1 and Core2.2 sites demonstrated the DR1 and DR2 accessory motifs of IRS specify all the required information to confer resolution selectivity to the reaction. Without DR1 and DR2, recombination at Core1.1 and Core2.2 occurred with all possible synapses, whereas recombination between directly repeated IRS1.1 and IRS2.2 derivatives only generated deletion products. For the pIRS1.2 × 2.1 reporter plasmid in which the orientation of one core site is reversed with respect to the other, the presence of DR1 and DR2 was found to inhibit recombination by preventing random synapsis in vitro. These data show that the accessory elements of the recombination complex do not only function in a topological filter mechanism to facilitate intramolecular synapsis between directly repeated IRSs, they also restrict the pairing of the recombination core sites, thereby increasing the overall efficiency of recombination by avoiding unproductive synapses.
In addition, recombination at wild-type IRS was found to follow a strictly ordered pathway in which the IR1-bound subunits mediate the first strand exchange to form the HJ intermediate and the IR2-bound subunits catalyse the second strand exchange to resolve the HJ into recombinant products. For the first time, this specific order of strand exchange was unambiguously demonstrated for a complete recombination reaction by trapping cleaved intermediates of both consecutive steps of the process. Analysis of the HJ intermediates formed at the other IRS variants showed that in all cases recombination was initiated by the TnpI subunits bound to the most distal core site motifs with respect to the accessory sequences, irrespective of the orientation of the core sites. In the absence of DR1 and DR2, strand exchange was only restricted by the proper alignment of the overlap regions within the core sites, producing alternative isoforms of the HJ intermediate. Together, these data demonstrate that the accessory sequences and proteins impose a specific arrangement of the core sites in the synaptic complex, selecting for one of the two active configurations of the recombinase tetramer to initiate recombination (Figure 1).
This may occur indirectly, by imparting a specific path to the core sites within the complex, or more directly by anchoring the catalytic TnpI subunits through specific protein–protein or protein–DNA interactions with the accessory elements. For the pIRS1.2 × 2.1 substrate, the specific architecture of the synapse brought the two sites in a normally non-productive parallel alignment; forcing the recombinases to exchange the ‘wrong’ strands and generating mismatched recombination intermediates. For forming an antiparallel synapse, one core site would have to rotate by 180° with respect to the other, which would necessarily disturb the organisation of proteins and DNA within the complex.
Accessory sequences and proteins were previously shown to determine the order of stand exchange catalysed by λ Int and E. coli Xer system (17,30–34). DNA integration and excision reactions mediated by λ Int are controlled by the formation of specific complexes in which positioning of the recombinase protomers to the core recombination sites is dictated by their interaction with flanking ‘arm’ sites through an extra DNA-binding domain located at the N terminus of the protein (35,36). Xer-mediated recombination at the chromosomal dif site is activated by the cell division protein FtsK (33,37), while recombination at plasmid resolution sites like cer (ColEI plasmid) or psi (pSC101) requires the assembly of a topologically defined synapse involving dedicated host proteins instead of extra-recombinase subunits as is the case for TnpI (18,38,39). Formation of this complex imposes a specific geometry to the recombinase core tetramer as we propose here for the TnpI/IRS system (34). However, in Xer recombination, an additional level of control on strand exchange is provided by the heteromeric structure of the XerCD recombinase (14,40). Recombination of a plasmid in which inversely oriented core sites were joined to directly repeated psi accessory sequences yielded inversion products with the topology of a three-noded knot (34) rather than aberrant strand-exchange products as we observed with pIRS1.2 × 2.1. This likely reflects weaker constraints for the positioning of the core sites in the Xer complex, and/or the requirement for complementary interactions between XerC and XerD to assemble the active XerCD tetramer.
The regulatory components of TnpI/IRS recombination complex direct strand-exchange catalysis by stabilizing reaction intermediates
An intriguing finding of this study is the strong accumulation of cleaved intermediates observed with the TnpI-H234L mutant at wild-type IRS (Figure 8). In the yeast recombinase Flp, the equivalent histidine residue (H305) was recently proposed to function as a general acid/base catalyst to assist the tyrosine nucleophile during strand exchange (41). Given the relatively high pKa of tyrosine (pKa ≈ 10), substitution of this residue is expected to have a more deleterious effect on DNA cleavage (during which it acts as a base to attract a proton from the tyrosine) than on DNA rejoining (where it acts as an acid by returning the proton to the leaving group). Indeed, mutation of the corresponding active site histidine of Cre (H289) was found to strongly reduce recombination activity due to a major drop (up to 1200-fold) in cleavage rate, as measured on suicide DNA substrates (41,42). Likewise, incubation of TnpI-H234L with linear substrates did not lead to covalent complexes accumulation, and recombination at the minimal IR1–IR2 core site was essentially undetectable (data not shown). Assuming that the H234L mutation does not alter the activity of other active-site residues of TnpI, we therefore propose that the presence of DR1–DR2 promoted a conformational change that stabilized cleaved intermediates produced by TnpI-H234L, thereby preventing their religation back to substrate. A similar conformational change is likely to take place to stabilize covalent complexes that are generated by the H305 mutants of Flp (28,29), although in this case recombination does not require accessory sequences and proteins.
In addition to be affected in recombination, the H289A mutant of Cre shows a reversed order of strand exchange compared to wild-type Cre and accumulates HJ intermediates (42). This indicates the His 289 residue of Cre is directly involved in the mechanism of DNA strand cleavage specificity that is dictated by the central region of the loxP recombination site. Although a similar mechanism may contribute to the directionality of the reaction catalysed by TnpI (see below), the H234L substitution did not change the order of strand exchange that was determined for wild-type TnpI based on HJ intermediates analysis. This, again, supports the view that specific positioning and activation of the catalytic TnpI subunits are primarily determined by the accessory elements of the TnpI/IRS complex.
The presence of adequately positioned DR1 and DR2 motifs was also found to stabilize the HJ intermediate of recombination. HJs accumulated to different extents with all full-length IRS derivatives examined, but were never observed with the minimal core-site substrates under standard conditions. Only small amounts of HJ products could be trapped when the reactions were performed in the presence of the hexapeptide inhibitor KWWCRW. This kind of peptide was shown to block HJ processing by altering the structure of the HJ-recombinase complex at the time of its formation (43). Stabilisation of strand-exchange intermediates either before or after DNA rejoining was also important to generate energetically unfavourable mismatched strand transfer products by recombination of misaligned sites on pIRS1.2 × 2.1.
Together, these data suggest a mechanism in which the regulatory components of the complex provide the required energy input to promote conformational changes in the catalytic core complex, thereby displacing individual equilibriums of the strand-exchange reaction towards product formation. These conformational changes are required to unwind DNA in the overlap regions of the recombination sites (Figure 1, steps II), to orient the cleaved 5′ OH ends for being resealed to the partner (steps III and IV), and then to mediate isomerization of the recombinase tetramer, switching TnpI activity within the complex (step V). A similar mechanism was recently proposed for λ Int based on biochemical and structural data corresponding to different stages of strand-exchange progression (12,36,44). In this case, the conformational switch that drives recombination is mediated by the interwoven interactions that link the core-bound recombinase subunits to the flanking arm sites.
Additional factors that modulate directionality: role of the core recombination site overlap region
For different members of the tyrosine recombinase family, the nucleotide sequence of the core recombination site central region was shown to influence the directionality of strand exchange, either by determining the direction of the initial bends in the core complex, thereby favouring one active configuration of the synapse over the other, or by acting on HJ isomerization (14,17,45–49). In the Cre/loxP system, asymmetry in the 8-bp central region of the loxP site dictates a specific order of strand exchange by Cre without requiring additional accessory sequences or proteins (17,47,49).
TnpI shows no preferential cleavage on linear DNA molecules and synthetic HJ substrates containing the IR1–IR2 sequence are resolved equally well by the IR1- and IR2-bound subunits (19) (data not shown). However, altering the IR1–IR2 spacer sequence was found to affect recombination in the context of supercoiled DNA substrates. Recombination at the symmetrical Core2.2 site was less efficient than recombination at Core1.1, and reactions performed with the pIRS2.2 × 2.2 substrate accumulated higher levels of HJ intermediates than observed with pIRS1.1 × 1.1 (Figure 5). More strikingly, reversing the normal order of strand exchange by inverting the orientation of the IR1–IR2 core site with respect to the accessory sequences was found to strongly inhibit recombination (Figure 4). This indicates that the IR1–IR2 spacer sequence is optimized to reinforce the action of the accessory elements by favouring HJ formation at the IR1 half sites and HJ resolution at the IR2 half sites.
Thus, the directionality of the DNA-strand-exchange reaction catalysed by different members of the tyrosine recombinase family appears to be controlled by a combination of different factors acting on common and highly conserved chemical steps of the recombination pathway. Further investigations are needed to fully understand how these factors affect the kinetics and thermodynamics of individual conformational changes taking place during recombination.
FUNDING
‘Fonds Spéciaux de la Recherche’ at UCL, the ‘Action de Recherche Concertée’ (grant number 01/06-268); ‘Fonds National de la Recherche Scientifique’ (‘Crédits aux Chercheurs’ grant numbers 1.5.177.01 and 1.5.190.03). Funding for open access charge: Personal grants.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to François-Xavier Barre, Marie Deghorain and Damien Dandoy for helpful discussions and comments on the manuscript, and to all the members of the Genetics Unit, past and present, for continuous support and encouragements.
REFERENCES
- 1.Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 2.Hallet B, Sherratt DJ. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 3.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 4.Van Duyne GD. A structural view of cre-loxp site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 5.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Rice PA. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu. Rev. Biophys. Biomol. Struct. 2003;32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- 7.Grainge I, Jayaram M. The integrase family of recombinase: organization and function of the active site. Mol. Microbiol. 1999;33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 8.Sherratt DJ, Wigley DB. Conserved themes but novel activities in recombinases and topoisomerases. Cell. 1998;93:149–152. doi: 10.1016/s0092-8674(00)81566-4. [DOI] [PubMed] [Google Scholar]
- 9.Aihara H, Huang WM, Ellenberger T. An interlocked dimer of the protelomerase TelK distorts DNA structure for the formation of hairpin telomeres. Mol. Cell. 2007;27:901–913. doi: 10.1016/j.molcel.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobryn K, Chaconas G. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell. 2002;9:195–201. doi: 10.1016/s1097-2765(01)00433-6. [DOI] [PubMed] [Google Scholar]
- 11.Nunes-Duby SE, Azaro MA, Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site-specific recombination. Curr. Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 12.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 14.Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- 15.Guo F, Gopaul DN, Van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 17.Lee L, Sadowski PD. Strand selection by the tyrosine recombinases. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:1–42. doi: 10.1016/S0079-6603(05)80001-7. [DOI] [PubMed] [Google Scholar]
- 18.Hallet B, Vanhooff V, Cornet F. DNA site-specific resolution systems. In: Funnell B, Phillips G, editors. Plasmid Biology. Washington, DC: ASM Press; 2004. pp. 145–179. [Google Scholar]
- 19.Vanhooff V, Galloy C, Agaisse H, Lereclus D, Revet B, Hallet B. Self-control in DNA site-specific recombination mediated by the tyrosine recombinase TnpI. Mol. Microbiol. 2006;60:617–629. doi: 10.1111/j.1365-2958.2006.05127.x. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 2009. [Google Scholar]
- 21.Cassell G, Klemm M, Pinilla C, Segall A. Dissection of bacteriophage lambda site-specific recombination using synthetic peptide combinatorial libraries. J. Mol. Biol. 2000;299:1193–1202. doi: 10.1006/jmbi.2000.3828. [DOI] [PubMed] [Google Scholar]
- 22.McCulloch R, Coggins LW, Colloms SD, Sherratt DJ. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turlan C, Loot C, Chandler M. IS911 partial transposition products and their processing by the Escherichia coli RecG helicase. Mol. Microbiol. 2004;53:1021–1033. doi: 10.1111/j.1365-2958.2004.04165.x. [DOI] [PubMed] [Google Scholar]
- 24.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Crisona NJ, Weinberg RL, Peter BJ, Sumners DW, Cozzarelli NR. The topological mechanism of phage lambda integrase. J. Mol. Biol. 1999;289:747–775. doi: 10.1006/jmbi.1999.2771. [DOI] [PubMed] [Google Scholar]
- 26.Grainge I, Buck D, Jayaram M. Geometry of site alignment during int family recombination: antiparallel synapsis by the Flp recombinase. J. Mol. Biol. 2000;298:749–764. doi: 10.1006/jmbi.2000.3679. [DOI] [PubMed] [Google Scholar]
- 27.Klemm M, Cheng C, Cassell G, Shuman S, Segall AM. Peptide inhibitors of DNA cleavage by tyrosine recombinases and topoisomerases. J. Mol. Biol. 2000;299:1203–1216. doi: 10.1006/jmbi.2000.3829. [DOI] [PubMed] [Google Scholar]
- 28.Parsons RL, Prasad PV, Harshey RM, Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol. Cell Biol. 1988;8:3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu XD, Sadowski PD. Cleavage-dependent ligation by the FLP recombinase. Characterization of a mutant FLP protein with an alteration in a catalytic amino acid. J. Biol. Chem. 1995;270:23044–23054. doi: 10.1074/jbc.270.39.23044. [DOI] [PubMed] [Google Scholar]
- 30.Franz B, Landy A. The Holliday junction intermediates of lambda integrative and excisive recombination respond differently to the bending proteins integration host factor and excisionase. EMBO J. 1995;14:397–406. doi: 10.1002/j.1460-2075.1995.tb07014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitts PA, Nash HA. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol. 1988;204:95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- 32.Nunes-Duby SE, Matsumoto L, Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987;50:779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- 33.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 34.Bregu M, Sherratt DJ, Colloms SD. Accessory factors determine the order of strand exchange in Xer recombination at psi. EMBO J. 2002;21:3888–3897. doi: 10.1093/emboj/cdf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azaro M, Landy A. Lambda integrase and the lambda int family. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 118–148. [Google Scholar]
- 36.Radman-Livaja M, Biswas T, Ellenberger T, Landy A, Aihara H. DNA arms do the legwork to ensure the directionality of lambda site-specific recombination. Curr. Opin. Struct. Biol. 2006;16:42–50. doi: 10.1016/j.sbi.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, Arciszewska LK. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol.Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colloms SD, Bath J, Sherratt DJ. Topological selectivity in Xer site-specific recombination. Cell. 1997;88:855–864. doi: 10.1016/s0092-8674(00)81931-5. [DOI] [PubMed] [Google Scholar]
- 39.Reijns M, Lu Y, Leach S, Colloms SD. Mutagenesis of PepA suggests a new model for the Xer/cer synaptic complex. Mol. Microbiol. 2005;57:927–941. doi: 10.1111/j.1365-2958.2005.04716.x. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira H, Sherratt D, Arciszewska L. Switching catalytic activity in the XerCD site-specific recombination machine. J. Mol. Biol. 2001;312:45–57. doi: 10.1006/jmbi.2001.4940. [DOI] [PubMed] [Google Scholar]
- 41.Whiteson KL, Chen Y, Chopra N, Raymond AC, Rice PA. Identification of a potential general acid/base in the reversible phosphoryl transfer reactions catalyzed by tyrosine recombinases: Flp H305. Chem. Biol. 2007;14:121–129. doi: 10.1016/j.chembiol.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelato KA, Martin SS, Baldwin EP. Reversed DNA strand cleavage specificity in initiation of Cre-LoxP recombination induced by the His289Ala active-site substitution. J. Mol. Biol. 2005;354:233–245. doi: 10.1016/j.jmb.2005.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh K, Lau CK, Guo F, Segall AM, Van Duyne GD. Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination. J. Biol. Chem. 2005;280:8290–8299. doi: 10.1074/jbc.M411668200. [DOI] [PubMed] [Google Scholar]
- 44.Radman-Livaja M, Shaw C, Azaro M, Biswas T, Ellenberger T, Landy A. Arm sequences contribute to the architecture and catalytic function of a lambda integrase-Holliday junction complex. Mol. Cell. 2003;11:783–794. doi: 10.1016/s1097-2765(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 45.Arciszewska LK, Baker RA, Hallet B, Sherratt DJ. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 2000;299:391–403. doi: 10.1006/jmbi.2000.3762. [DOI] [PubMed] [Google Scholar]
- 46.Azaro MA, Landy A. The isomeric preference of Holliday junctions influences resolution bias by lambda integrase. EMBO J. 1997;16:3744–3755. doi: 10.1093/emboj/16.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh K, Lau CK, Gupta K, Van Duyne GD. Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat. Chem. Biol. 2005;1:275–282. doi: 10.1038/nchembio733. [DOI] [PubMed] [Google Scholar]
- 48.Lee L, Sadowski PD. Directional resolution of synthetic holliday structures by the Cre recombinase. J. Biol. Chem. 2001;276:31092–31098. doi: 10.1074/jbc.M103739200. [DOI] [PubMed] [Google Scholar]
- 49.Lee L, Sadowski PD. Sequence of the loxP site determines the order of strand exchange by the Cre recombinase. J. Mol. Biol. 2003;326:397–412. doi: 10.1016/s0022-2836(02)01429-8. [DOI] [PubMed] [Google Scholar]