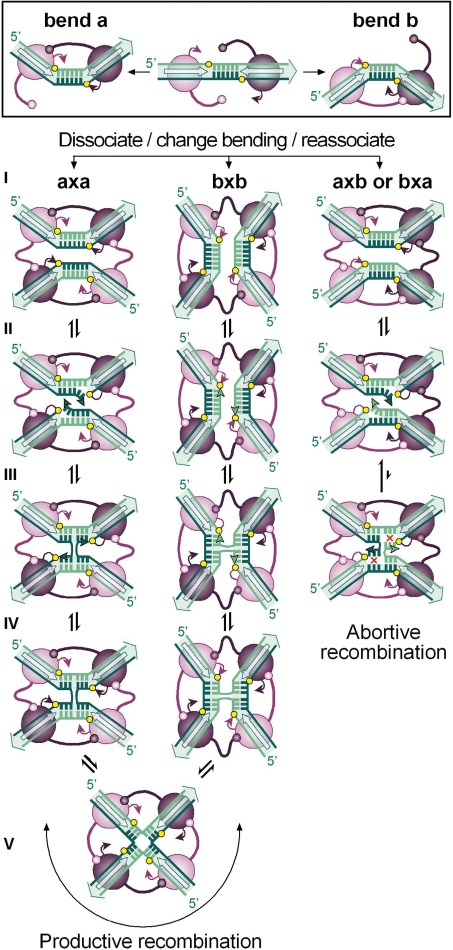
Figure 1.
The strand-exchange reaction catalysed by tyrosine recombinases. Binding of a pair of recombinase molecules (purple ovals) onto the core recombination site recognition elements (open arrows) may bend DNA to opposite directions (a or b) exposing one or the other strand for catalysis (coloured dark and light green, respectively). In all cases studied, activity of the recombinase subunits is controlled by allosteric interactions in which the active subunit donates its C-terminus (represented by a stem-and-ball extension) to its neighbour. During synaptic complex formation (I), both recombination sites may pair with an antiparallel alignment if they are in the same bent configuration (a × a or b × b) or with a parallel alignment if they are bent in opposite directions (a × b or b × a). In all cases, this leads to the formation of a recombinase tetramer in which diagonally opposed subunits are activated, while the other pair of subunits is inactive. Antiparallel pairing of the recombination sites proceeds to strand exchange through a sequence of reversible steps during which the exposed strands are cleaved (II), exchanged (III) and then rejoined (IV) to the partner strand to form a HJ. Isomerization of the HJ (V) inactivates one pair of recombinase subunits and activates the other pair of subunits to exchange the second pair of strands. For each strand-exchange reaction, the recombinase catalytic tyrosine (purple arrowhead) attacks the adjacent sessile phosphate (yellow circle) to form a phosphotyrosyl bond that is in turn attacked by the 5′OH end of the cleaved DNA strand (green arrowhead). Note that recombination that initiates with one configuration of the synapse terminates with the core sites in the opposite configuration and reciprocally. Parallel pairing of the recombination sites generally leads to abortive recombination due to the lack base pairs complementarity (red crosses) in the overlap region (step III).