Figure 8.
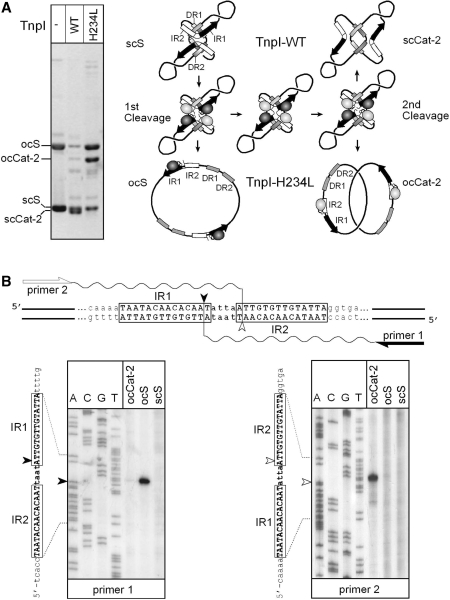
Ordered DNA cleavages during TnpI-catalysed sequential strand exchanges (A) TnpI and TnpI-H234L recombination pathways. A reporter plasmid carrying directly repeated IRSs was incubated with wild-type TnpI (TnpI-WT) or TnpI-H234L, and the reactions were run uncut on a 0.8% agarose gel. Both proteins assembled onto the supercoiled substrate (scS) to form the TnpI/IRS-specific synapse. For clarity, only one of both possible topological organizations of the synaptic complex is shown (19). The IR1–IR2 core sites and the DR1 and DR2 accessory motifs are represented by black/white arrows and shaded boxes, respectively. The core TnpI subunits bound to IR1 and IR2 half sites are shown as dark and light shaded ovals, respectively. Sequential strand-exchange reactions catalysed by TnpI-WT leads to fully supercoiled two-noded catenane products (scCat-2). In the presence of TnpI-H234L, DNA rejoining was partially inhibited leading to the formation of open circular forms of the substrate (ocS) and catenane product (ocCat-2). (B) Mapping of TnpI-H234L DNA cleavages. Gel-purified products arising from the first (ocS) and second (ocCat-2) DNA cleavage reactions were used as a template in primer extension analysis to determine whether cleavage occurred at IR1 (primer 1) or IR2 (primer 2). Extension products were analysed on a 6% sequencing gel alongside of a DNA sequencing reaction performed with the same primers. Extensions performed with unreacted substrate (scS) were used as a control in the experiment. IR1 and IR2 cleavage positions are shown by a filled and open arrowhead, respectively. Note that the thermostable DNA polymerase used in the experiment added an extra dA nucleotide at the 3′ end of the extension products.