Figure 6.
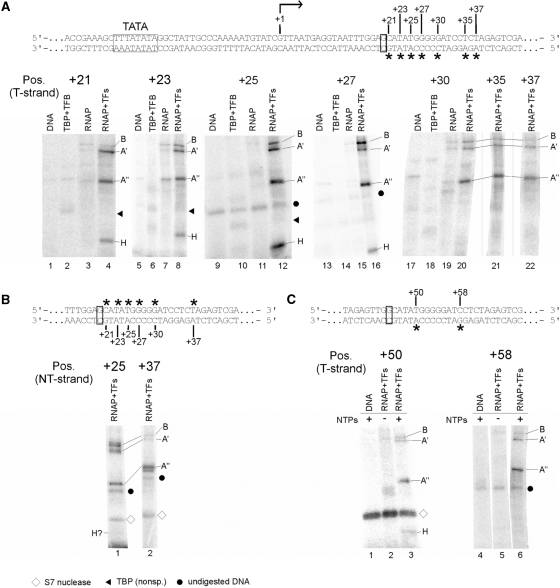
Mapping of RNAP subunits cross-linked to the gdh promoter DNA in stalled elongation complexes. (A) Processivity of ECs stalled at +20 is not affected by TFS. Increasing amounts of purified TFS were added to stalled ECs (lanes 4–9) and complexes were chased with a complete set of NTP after incubation in the absence (lane 3) and presence of TFS (lane 8). The amount of non-chaseable RNA in complexes is insignificant in both reactions and the amount of synthesized run-off transcript ∼ the same indicating that the stalled complexes were not backtracked in the absence of TFS. As expected, the 21-nt RNA most likely generated by misincorporation of an unpaired nt was removed upon TFS treatment. (B) Photocross-linking of proteins in a ternary complex stalled at position +20. The gdh promoter template spans bp −39 to +66 relative to the transcription start site (bent arrow). TATA box and the stalling site +20 are boxed. Locations of photoactivatable labels (4-azidophenacyl bromide coupled to a phosphorothioate modification in the DNA backbone) are indicated by asterisks. RNAP purified from Pyrococcus cells was used to stall elongation complexes at +20 as described earlier (6,27). Cross-linked subunits of the stalled complexes were analyzed via 4–20% SDS-PAGE. The position of the photoactive cross-linking site in the transcribed strand is indicated above the gels. Proteins present in the individual reactions are specified on top of the gels. Radiolabeled RNAP subunits were identified by their relative electrophoretic mobility and are indicated at the right-hand side of the gels. Dots indicate non-specific signals derived from undigested DNA, whereas triangles indicate non-specific TBP cross-links. (C) Cross-linking of RNAP subunits in stalled elongation complexes to the non-transcribed strand. Positions analyzed are indicated by asterisks above the sequence. No cross-linking of H was detectable at any of the tested positions except of position +25, where a very faint band suggests weak cross-linking of H. To exemplify this, positions +25 and +37 are shown. (D) H interacts with DNA close to the active center also in late elongation complexes. Mature ECs, stalled at position +45 were cross-linked to position +50 and +58 on the T-strand as described in (B). The diamond marks auto-radiolabeled S7 nuclease. Part of the DNA sequence is shown at the top of the panel.