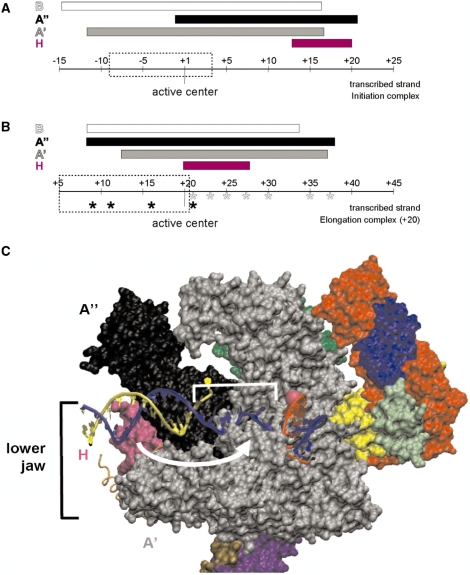
Figure 9.
Schematic summary of the repositioning of H and A′′ in elongation complexes. The range of protein/DNA interaction of RNAP subunits H (magenta), A′ (black), A′ (gray) and B (white) is symbolized by bars. The position of the RNAP’s active center is highlighted and the extension of the transcription bubble (27) is marked by dashed boxes. (A) Interactions of archaeal RNAP subunits with the transcribed DNA-strand in the PIC. H marks the far downstream end of the complex (based on the results published by ref. 17). (B) Schematic representation of cross-links of archaeal RNAP in a ternary complex stalled at +20. Cross-linking from position +9 to +21 was analyzed previously (6), and those derivatized sites are labeled by black asterisks. Cross-linkers from position +21 to +37 from this study are indicated with gray asterisks. Note that H is localized in close proximity to the active center and that the upstream boundary of A′′ is extended by ∼10 nt in stalled ECs. (C) Model for the path of DNA relative to RNAP in the archaeal elongation complex. The S. shibatae RNAP structure (PDB ID: 2WAQ; 13) was aligned with eukaryotic RNAP II in an EC (PDB ID: 2E2H; 35) using C-alpha coordinate ‘Iterative Magic Fit’ from Swiss PDBViewer 4.01 (54). R.M.S. deviation of the aligned structures was 1.55 Å for 1786 atoms. The RNAP II EC DNA and RNA coordinates were then merged with the archaeal RNAP structure coordinates and rendered concurrently using VMD 1.8.6 (54), revealing a good fit with few clashes. RNAP subunit colors are as used in (9), except for subunits G (darker green, obscured) and Rpo13 (orange trace) that were not part of the structure solved by (9). Subunit B was removed to allow visualization of the nucleic acids. The DNA transcribed and non-transcribed strands are blue and yellow, respectively, and the RNA is red. The active site is indicated by a pink sphere, and represents the +1 position in the EC. +1 to +10 of the T strand is bracketed, and the white arrow suggests the conformational change that subunit H (in magenta) would need to make to be cross-linked by aryl azide derivatizations within this region.