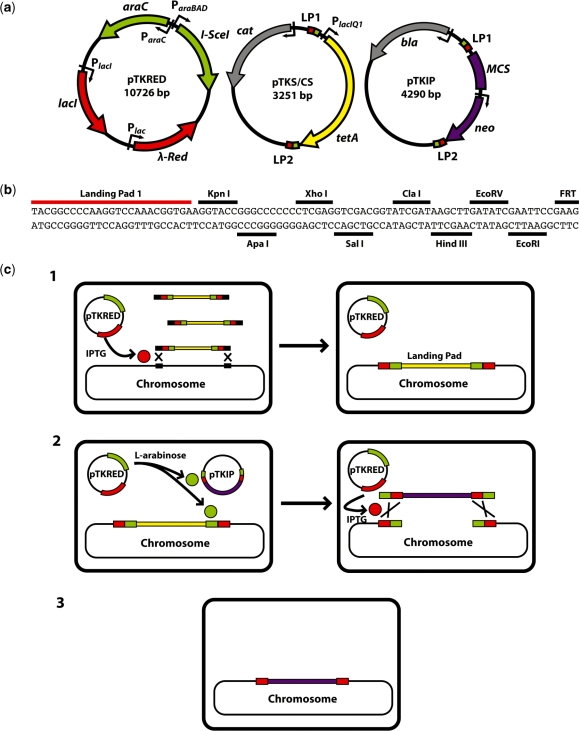
Figure 2.
(a) Plasmids used in the integration protocol. The sequence size given is for pTKIP-neo; neo is exactly replaced with various antibiotic resistance genes for alternate versions of pTKIP. Small green boxes are I-SceI restriction sites; landing pad regions 1 and 2 are small red boxes labeled LP1 and LP2 respectively. (b) Annotated sequence of the pTKIP MCS showing LP1, available restriction sites, and the first four bases of the adjacent FRT site. (c) Strategy for large construct chromosomal integration. Step 1: the host strain is transformed with the helper plasmid pTKRED, bearing I-SceI endonuclease (green) and λ-Red (red). Linear landing pad fragments (yellow) are integrated into the chromosome at the desired location (black squares) when λ-Red expression is induced by IPTG. Step 2: the host strain is transformed with pTKIP bearing the fragment (purple) to be inserted into the landing pad. I-SceI expression is induced via the addition of l-arabinose, and the I-SceI recognition sites (green) in the donor plasmid and chromosome are cleaved. Integration of the fragment is facilitated by IPTG-induced λ-Red expression. Step 3: pTKRED is cured by growth at 42°C and screening against spectinomycin resistance.