Abstract
Insight into the pathogenesis of neurodegenerative disorders requires accurately categorized postmortem human brain tissue. This article introduces electronic tissue tracking and management as implemented at New York Brain Bank (NYBB) through processing of the brain at fresh state and storing standardized frozen samples. NYBB tissue tracking uses a relational database to co-register a bar coded, unique sample identifier to unique coordinates in the three-dimensional freezer space, allowing immediate retrieval of stored samples without further dissection. In the 5 years since the inception of NYBB (2002-2007) 560 brains (63,252 fresh frozen samples) were processed and as of 11/2007, 54,242 samples are stored seven freezers occupying 81% of maximum capacity of NYBB. Within the same time period, 1,094 requests were processed and 9,096 samples were disbursed with an average turnaround time of five working days. The NYBB system of brain banking has the following key advantages: (1) The dissection of the brain and the harvest of samples at the fresh state improve their anatomic specificity and quality; (2) samples are ready for immediate disbursement once categorized diagnostically, reducing the time between the receipt of request and disbursement of samples; (3) the methods prevent thaw-refreeze cycles and carving out of regions of interest from frozen tissue, which is cumbersome and deleterious to the both samples and source brains; (4) accurate quantitative data on stored samples according to anatomical regions and distributive diagnosis guides future sample collection and fosters effective use of limited resources.
Keywords: Brain banking, Electronic tracking, Human tissue, Neurodegenerative disorders, Tissue banking
Introduction
Insight into the pathogenesis of neurodegenerative disorders exclusively occurring in humans requires accurately categorized postmortem human brains, including brains of healthy control subjects (Stopa and Bird 1989; Ravid et al. 1995), to unravel the mechanisms underlying dementia, movement disorders or psychiatric diseases (Grinberg et al. 2007). The infrastructure and logistics of providing postmortem human central nervous system tissue is a rapidly evolving field that increasingly receives attention and funding (Ravid et al. 1992; Alafuzoff and Winblad 1993; Grinberg et al. 2007; Schmitt et al. 2007).
A central aspect of this endeavor at New York Brain Bank (NYBB) has been changing the paradigm from freezing the donated brain en bloc pending future dissection to processing of the brain immediately at fresh state and storing frozen samples of standardized dimensions and contents, i.e. Standardized Brain Block (SBB) and Standardized Cortex/White matter Vial (SCxV/SWmV) (Vonsattel et al. 1995, 2007).
This approach enhances anatomic accuracy of dissection, which optimizes the reliability of the samples, especially when comparing pathologic findings across different study groups synchronously or metachronously, as new molecular targets become available. Storing standardized, anatomically and pathologically well categorized samples of narrowly confined dimensions, which can be tightly packed into defined storage compartments with little “dead space” and electronically tracked, enhances the usage of freezer cabinets as compared to en bloc frozen brains, half brains, tissue slabs or blocks with variable dimensions. This procedure combined with the capability of electronically monitoring freezer compartments made available by sample disbursement, which can be replenished with newly procured samples, improves the efficiency of the freezer space by 40%. Locating standardized tissue samples in freezer compartments with unique coordinates reduces retrieval time, thus decreasing temperature fluctuations (harmful to tissue and freezers), labor costs (time spent searching samples) and turnaround time.
To manage collection, storage and distribution of tens of thousands of standardized tissue samples, each sample and freezer storage compartment requires a unique identifier and, subsequently, both need to be co-registered in a database, further referred to as NYBB tracking system. Bar coding these identifiers, shortens data entry time and secures data accuracy. The NYBB tracking system links storage information to the donor's demographical information and neuropathologic characteristics, thus allowing the allocation of tissue based on the requestor's stated parameters.
Materials and methods
Computers and printers
The NYBB tracking system is hosted on an Apple (Apple, Inc., Cupertino, CA, USA) Power Mac G4 (FW800) with 2 GB PC2700 (333 MHz) DDR SDRAM, 2 80 GB Ultra ATA/100 (7,200 rpm) hard drives and a SuperDrive (DVD-R/CD-RW), running Mac Os X Server (10.2.8). The hard drives are synchronized automatically and, additionally, data of the last five business days are kept under lock in a different room on Compact Disc ReWritables (CDRWs). An Apple iMac G3 (PowerPC), 256 MB SDRAM, Mac Os X (10.2.8), is located in a dedicated “brain cutting room” to create a unique database record and bar code label (Fig. 1) for every sample as it is obtained during the dissection of usually one half of any fresh brain donated for research. A computer of similar specifications is installed in the “freezer room” to scan tissue samples and co-register them to a compartment with unique coordinates in the freezer space (see below). Each computer (brain cutting and freezer rooms) is connected to a Dymo Label Writer 310 printer (Dymo, Stamford, CT, USA) to print labels for samples (Figs. 1, 2) and boxes/freezers (Fig. 3, panels c and b). Several computers of a similar configuration are used for: data entry (administrative assistant), request preparation and handling (tissue processing assistant) and data evaluation (research fellow). All computers are connected to the university LAN/T1 (local area network) and protected from unauthorized access.
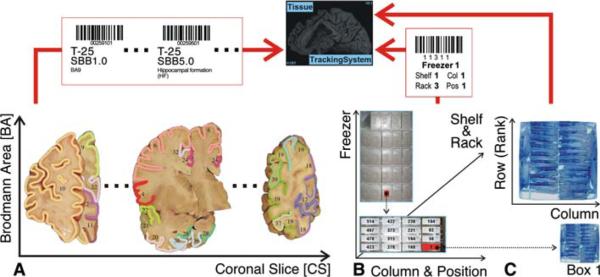
Fig. 1.
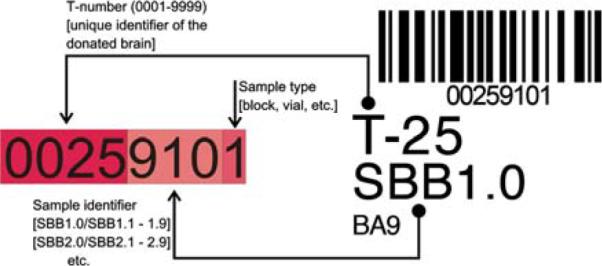
NYBB bar code label (right side). The bar code contains the unique identifier of the stored NYBB sample; here, tissue block SBB1.0 (Standardized Brain Block 1 [BA9]) of brain T-25 (unique identifier of the donated brain, which is assigned at receipt of the specimen) is depicted. The sample type is 1, which denotes a tissue block. The label is placed within the sample bag, to ensure that the identifier will not be accidentally removed. The bar code is scanned through the sample bag
Fig. 2.
NYBB bar code labels. (a) Self adhesive labels with the bar coded unique sample identifier and printed sample information are placed in the sample bags with the frozen tissue blocks. (b) NYBB labels are attached to the outside of the vials containing pulverized brain parenchyma. Samples are scanned upon placement in the freezers and retrieval for disbursement
Fig. 3.
Schematic of NYBB Tracking System storage information. The relational database (NYBB Tracking System) correlates a bar coded, unique sample identifier (panel a)with a unique set of bar coded coordinates in the 3D freezer space (panel b). Both the unique identifier and the freezer coordinates are bar coded to minimize the incidence of data entry error. The box position (panel c) is assigned by the computer and depends on the availability of the open (not filled) boxes as well as the sampled region (optional)
Database
FileMaker Pro 6.0 (FileMaker, Inc., Santa Clara, CA, USA), a relational database, was used to implement NYBB tracking system through customized scripts in a generic scripting language of FileMaker Pro 6.0, which facilitate data entry, i.e., neuropathologic diagnosis, tissue categorization, sample storage and request management. NYBB tracking system is hosted on a server (see above) using FileMaker Server 5.5 (FileMaker, Inc., Santa Clara, CA, USA). Access to the server is password restricted through FileMaker Pro 6.0 and file privileges for the database are tailored to the tasks for each member of NYBB to prevent accidental deletion/alteration of data and protect the identity of the donors.
Bar coded sample identifier
The bar coded unique sample identifier (Fig. 1) is composed of a four digit case number (T-number, assigned to the donated brain as it is received by NYBB staff), a three digit numerical identifier (encoding the anatomical region of the brain from which the sample is obtained) and a single digit sample type (coronal slice, tissue block [SBB] or vial [SCxV, SWmV] of pulverized brain tissue). The details of the regions encoded by the numerical identifier (SBB; SCxV; SWmV) are described in greater detail elsewhere (Vonsattel et al. 2007) and are available through the NYBB Web pages (http://www.nybrainbank.org). Every sample is frozen in liquid nitrogen vapor, according to the protocol described elsewhere (Vonsattel et al. 2007) and immediately labeled with a unique bar code tag for tracking purposes. The “Code 128 Barcode Font Package” (IDAutomation, Tampa, FL, USA) is used to generate a bar code through a third party plug-in (IDAutomation, Tampa, FL, USA) for FileMaker Pro 6.0. The bar code labels are printed on standard labels (1″ × 1″, 750 labels/roll, catalog no. 30332) supplied Dymo, Stamford, CT, USA. These self-adhesive labels are either inserted in the transparent Fisherbrand® Bitran specimen bag (Fisher Scientific, Pittsburgh, PA, USA; 2 × 4 in.; catalog no. 19-240093) with the frozen tissue blocks (Fig. 2, panel a) or tightly applied to the outside of the vials (Wheaton® Cryules—Fisher Scientific, Pittsburgh, PA, USA; catalog no. 03-341-18) containing frozen, pulverized brain parenchyma (Fig. 2, panel b). All bar codes are read either with a cordless scanner (Worth Data, Santa Cruz, CA, USA; one-way radiofrequency CCD base station, catalog no. B62; oneway radiofrequency CCD scanner, catalog no. LI101-RF) or a VoyagerCG® scanner (Metrologic, Blackwood, NJ, USA; catalog no. MS9540), which are both connected to the freezer room computer (see above).
Compartmentalization of freezers
The bar coded samples are stored in Thermo Scientific Revco® Ultralow (-80°C) freezers (Thermo Fisher Scientific, Inc., Waltham, MA, USA; model nos. ULT2186-9-A36/-A12/-A30), containing five shelves with separate latch doors. Each shelf accommodates five stainless-steel inventory freezer racks (Fisher Scientific, Pittsburgh, PA USA; for 2 in.-high boxes [see below], catalog no. 11-678-29E), except the top shelf, which holds only four racks, due to structural space constraints. Each rack contains sixteen compartments for fiberboard storage boxes (Fisher Scientific, Pittsburgh, PA, USA; 2 in. high, catalog no. 11-678-24A) in four columns of four compartments each (4 × 4 = 16). The total number of compartments per freezer is 384 (24 racks with five racks on four shelves [5 × 4 = 20] and four racks on the top shelf = 24; and 24 × 16 compartment per rack = 384).
Each freezer compartment can be assigned unique coordinates (freezer, shelf and rack number with column and row position within the rack; Fig. 3, panel b), which are printed on a bar code tag applied to the rack wall of each compartment. Using these bar code tags, box placement into and retrieval from compartments can be confirmed electronically with minimal risk of data entry errors. Any box can be assigned a new compartment by scanning the compartment tag as the box is placed. Freezer compartmentalization also allows “real-time” tracking storage capacity of NYBB freezers, guiding the extent of future brain dissections. In the event of freezer malfunction all racks are transferred into a “backup” freezer, conserving the topography of the freezer space, and storage and disbursement can proceed uninterrupted while the malfunctioning freezer is serviced without the need to update the computer records.
Compartmentalization of boxes
The compartmentalization is extended by assigning unique coordinates to positions within the SBB/SCxV/SWmV freezer boxes (see above), to accommodate frozen tissue blocks (Fig. 4) and vials (Fig. 5). Thus, unassigned box position, which have become available through sample distribution, can be tracked and reassigned to new samples, further decreasing the amount of unused freezer space.
Fig. 4.
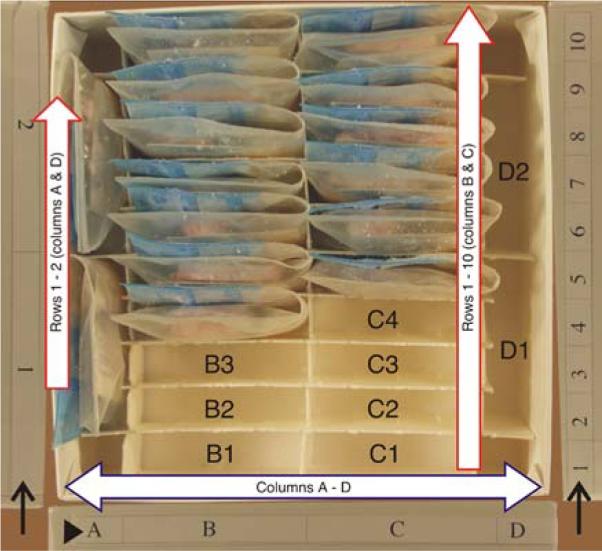
NYBB tissue block freezer box (SBB-box). The freezer box for tissue blocks holds 24 samples, stored in four columns with two rows (columns A and D) or 10 rows (columns B and C). The coordinate grids are printed on the outside of the box for columns (arrowhead) and for rows (arrow) to make retrieval of tissue easier. As samples are added and removed the bar code tags are scanned to confirm accurate placement and reduce errors. New samples are placed in the remaining spaces; the next slot to be filled in this box would be “B1”
Fig. 5.
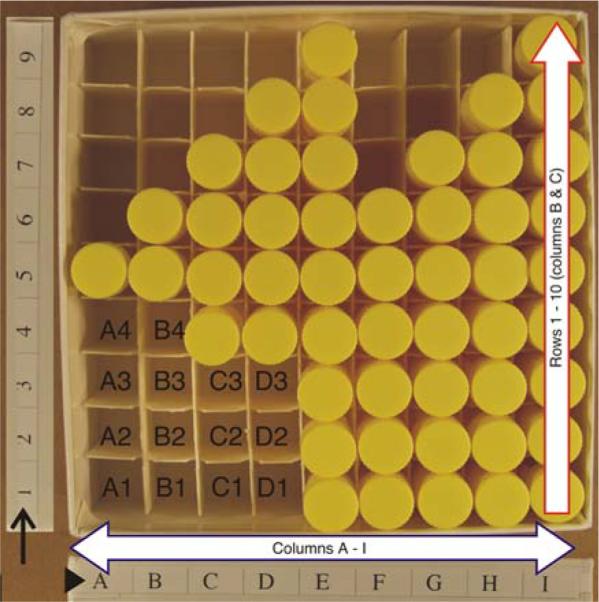
NYBB tissue vial freezer box (SCxV-/SWmV-box). The freezer box for tissue vials holds 81 samples, stored in nine columns with nine rows each. The coordinate grids are printed on the outside of the box for columns (arrowhead) and for rows (arrow) to make retrieval of tissue easier. As samples are added and removed the bar code tags are scanned to confirm accurate placement and reduce errors. New samples are placed in the remaining spaces; the next slot to be filled in this box would be “A1”
An SBB box has 24 positions (for storing fresh frozen blocks), referenced in a rectangular layout as A1-2, B1-10, C1-10 and D1-2, which is obtained by modifying fiberboard box dividers (Fisher Scientific, Pittsburgh, PA, USA; grid 7/16 in., 100 cell, catalog no. 11-678-24C) by removing vertical dividers 2-4 and 6-8, and curtailing the horizontal dividers 2-4 and 6-8 by one grid length at either end to accommodate 24 frozen tissue blocks (Fig. 4). Only 15 positions are occupied in the box depicted in Fig. 4. The position on the lower left (with the coordinates of B1) is not occupied and will be used for placement of the next sample by NYBB tracking system. An SCxV/SWmV box (for storing vials containing frozen pulverized brain aliquots) has 81 positions, referenced in a chessboard grid from A1-9 to I1-9 (Fig. 5), which is obtained with unaltered fiberboard box dividers (Fisher Scientific, Pittsburgh, PA, USA; grid 1/4 in., 81 cells, catalog no. 13-989-218). These boxes are identical except for the internal compartmentalization, which is customized either for fresh frozen blocks or for the vials containing pulverized frozen brain parenchyma.
The box layout is indicated on each box with self-adhesive labels indicating the row (ranks) and columns (Figs. 4, 5, arrows and arrowheads), to ease the location of a position according to the coordinates supplied by NYBB tracking system. At NYBB, a SBB or SCxV-/SWmV box contains blocks or vials from the same anatomical area of up to 24 or 81 different source brains.
In summary, NYBB tracking system co-registers a bar coded, unique sample identifier to unique coordinates in the three-dimensional freezer space, spanning an unlimited number of freezers and compartmentalizing the available space to the level of individual SBB or SCxV/SWmV (Fig. 3).
Tissue requests
Standardized tissue requests are retrieved through a WWW form, accessible only to investigators who have supplied an IRB approval and signed a material transfer agreement, available through the NYBB Web pages. A customized script, using a cross platform programming language, PERL (Practical Extraction and Report Language; http://www.perl.org/), queries contact information and request specifications in an interactive process, channels the investigator to the available regions and sends the request electronically to NYBB. Upon receipt, NYBB tracking system matches the requested tissue with the neuropathologically characterized, available tissue samples in a semiautomated process. Once the available samples have been assigned to an investigator, they are retrieved, coregistered with the tissue request through their unique bar code tags and disbursed. A description of the disbursed samples containing diagnosis, sex, age, ethnicity of the donor and the sampled region is automatically generated and accompanies the shipment.
It is important to note that NYBB tracking system greatly facilitates the quick access to a pool of “source brains” for fulfilling requests. The process of assembling and sampling source brains is among the most time consuming and tedious steps in the completion of a request with traditional storage systems.
Inventory
The following storage and processing information is available through automated database queries: (1) the number of processed brains and samples in a user defined time interval; (2) the number of currently stored samples; (3) the current freezer storage capacity and the percentage of freezer space used; and (4) the number of disbursed samples in a user defined time interval.
Results
Setup and transition period
With the hardware in place, installing and launching NYBB tracking system was accomplished within 1 week, including training of all staff and troubleshooting installation problems, e.g., updating printer drivers, adjusting page margins and customizing printouts. Before launching NYBB tracking system approximately 3,500 samples were accrued, using the NYBB protocol (Vonsattel et al. 1995, 2007), and these had to be refitted with appropriate bar code labels, scanned and placed in storage freezers. This process was accomplished within 3 months and required three staff members dedicating approximately 80% of their workday.
Processed brains
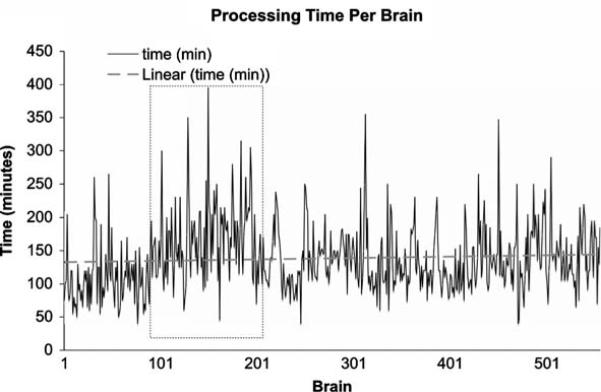
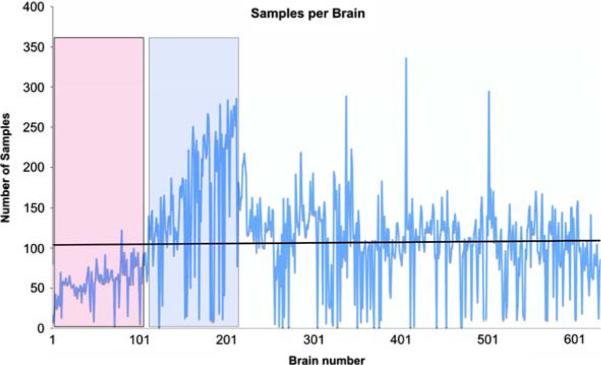
In the 5 years since the inception of NYBB (2002-2007) 560 brains (63,252 samples) were processed according to the previously published protocol (Vonsattel et al. 1995, 2007), using NYBB tracking system. Average processing time per brain was 2 h and 22 min, ranging from 40 min to 6 h and 35 min, depending mainly on the extent of dissection, on whether smears were generated, or on whether the opportunity of a dissection was used to demonstrate the protocol to interested neuroscientists or residents, guided by intake diagnosis and tissue quality. Despite the 3-fold increase in the number of disbursed samples from 923 in 2003, the first fully operational year of NYBB, to 2,865 in 2006 (see below and Table 2), the processing time per brain has remained relatively stable (Fig. 6), indicating that the standardized samples cover approximately 95% of requests, thus obviating the need for collecting customized samples. Slightly longer processing times were seen for brains 100-200 (Fig. 6, box with dotted outline), coinciding with the acquisition of a “capital of samples” in 2003, as NYBB became fully operational. Occasional spikes in processing time indicate rare disease entities or control brains, which are extensively sampled. Control brains are defined as brains without a known neurological or psychiatric disorder and without abnormality on gross examination during the dissection, and thereafter as per microscopic examination of the standardized series of blocks obtained from each brain banked. Coinciding with the increase in processing times, the number of harvested samples has peaked in 2003 (21,148 samples, Fig. 7), mainly through an increase in the number of samples obtained from the Alzheimer Disease Research Center (ADRC) at Columbia University. As the freezer capacity has reached 81% of the maximum capacity (see below), the acquisition of further samples has been slowed as is depicted in Fig. 7, without decrease in the number of samples harvested per brain (Fig. 8). Indeed, the number of processed samples per brain has been remarkably stable with a phase of slow increase at the inception of NYBB (Fig. 8, red box) and an accelerated phase to increase the “capital of samples” (Fig. 8, blue box), which coincided with the peak in processing times, described above (Fig. 6).
Table 2.
Number of request and disbursed samples per year
| Year | Requests | Blocks | Vials | Slices | Total |
|---|---|---|---|---|---|
| 2002 | 39 | 6 | 181 | 0 | 187 |
| 2003 | 184 | 430 | 477 | 16 | 923 |
| 2004 | 288 | 839 | 793 | 20 | 1,652 |
| 2005 | 212 | 1,053 | 695 | 12 | 1,760 |
| 2006 | 209 | 1,390 | 1,462 | 13 | 2,865 |
| 2007 | 162 | 752 | 950 | 7 | 1,709 |
| Total | 1,094 | 4,470 | 4,558 | 68 | 9,096 |
From 2002 to 2007, 1,094 requests have been processed with disbursement of a total of 9,096 samples. The number of disbursed samples per year has been increasing steadily from 2002 to 2006; however, no increase in the number of disbursed samples is projected for 2007
Fig. 6.
Processing times for NYBB brains. The processing time has remained fairly constant over the 558 brains processed. The time between brain 100 and brain 200 (box with dotted outlines) has slightly increased processing times, because dissections were more extensive to build a capital of tissue samples and has since returned to levels seen at the inception of NYBB
Fig. 7.
Processed samples of NYBB per year. The number of harvested samples peaked in 2003, when NYBB was building a capital of samples. This peak coincides with the increased processing times. ADRC, Alzheimer Disease Research Center; MD, movement disorders; WHICAP, Washington Heights and Inwood Columbia University Ageing Project; CPMC, Columbia Presbyterian Medical Center
Fig. 8.
Harvested samples per brain. The number of harvested samples per brain has remained fairly constant. The red box shows a gradual increase of the number of samples per brain as NYBB became operational and the blue box shows a peak in the number of samples as NYBB was acquiring a capital of samples. Occasional peaks in the number of samples per brain indicate rare diagnostic entities or control brains, which are extensively sampled
Stored samples
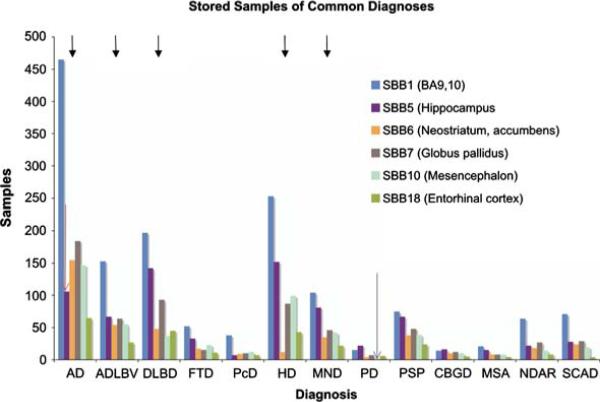
The available distributive diagnoses are shown in Table 1. Alzheimer disease is the most frequent diagnosis (n = 133), followed by Huntington Disease (n = 75) and Diffuse Lewy Body Disease (n = 56), reflecting the main specimen sources ADRC, WHICAP and Movement Disorders (Fig. 7). As of 11/2007, 54,242 fresh frozen samples are stored seven freezers in 2,165 boxes, occupying 81% of maximum capacity of NYBB. When storage levels for representative regions of common distributive diagnoses (Fig. 9, black arrows) are examined, patterns emerge, which will guide future sample collection at the NYBB: Area BA9 of Alzheimer disease (AD) brains are overrepresented while storage levels of the hippocampus (SBB5 [body], Fig. 9, red arrow) indicate that most investigations using postmortem human brain samples on AD are currently focused on the hippocampal formation. Similarly, levels of the mesencephalon (SBB10) of Parkinson disease brains (PD) indicate that most requestors favor this block for their studies (Fig. 9, blue arrow).
Table 1.
Number of NYBB brains in major diagnostic categories
| Diagnosis | Brains |
|---|---|
| AD | 131 |
| ADLBV | 44 |
| DLBD | 56 |
| TOD | 13 |
| FTD | 16 |
| PcD | 6 |
| VD | 3 |
| SCAD | 17 |
| HD | 75 |
| MND | 33 |
| PSP | 27 |
| PD | 9 |
| CBGD | 6 |
| MSA | 6 |
| DRPLA | 1 |
| CJD | 4 |
| CTRL | 42 |
| CIRC | 6 |
| Other | 155 |
| Total | 650 |
The majority of brains have a distributive diagnosis of Alzheimer Disease (AD, n = 133), followed by Huntington Disease (HD, n = 75) and Diffuse Lewy Body Disease (DLBD, n = 55). ADLBV, Alzheimer Disease Lewy body variant; TOD, tangle only dementia; FTD, frontotemporal dementia; PcD, Pick disease; VD, vascular dementia; SCAD, Senile changes of the Alzheimer type; MND, motor neuron disease; PSP, progressive supranuclear palsy; PD, Parkinson disease; CBGD, corticobasal ganglionic degeneration; MSA, multisystem atrophy; DRLPA, dentatorubral pallidolyusian atrophy; CJD, Creutzfeldt Jacob disease; CTRL, control brain; CRIC, circulatory abnormalities without dementia
Fig. 9.
Representative SBB for selected diagnostic categories. Area BA9 of Alzheimer disease (AD) brains are overrepresented while storage levels of the hippocampus (SBB5, red arrow) indicate that most research on AD is currently done on this block. Similarly, levels of the mesencephalon (SBB10) of Parkinson disease brains (PD) indicate that most requestors favor this block for their studies (blue arrow). Future sample collection at the NYBB will be guided to offset these imbalances to maximize use of precious resources
Specimen disbursement
In a 5-year period (2002-2007), 1,094 requests have been processed with disbursement of a total of 9,096 fresh frozen samples (Table 2) and an average turnaround time of five working days, provided that samples were available at receipt of the request and no prospective sampling was required (majority of cases). The number of disbursed samples per year has been increasing steadily from 2002 to 2006 (Table 2); however, no increase in the number of disbursed samples is projected for 2007. It is of interest to note that of the total of fresh frozen samples disbursed, 50% consists of vials of frozen pulverized tissue and 50% of tissue blocks.
The maximum number of samples disbursed from a single brain in the 5-year period from 2002 to 2007 was 100 samples (brain T-638, without a recognized diagnostic abnormality) in a total of 36 different requests. Had this brain been frozen en bloc, it would have been necessary to subject the brain to 36 thaw-refreeze cycles in order to carve out regions of interest for the various requests, thus jeopardizing tissue quality and limiting anatomical accuracy for subsequent requests.
Tracking non-standardized samples
While NYBB tracking system has been devised to work optimally with standardized brain samples, it retained considerable flexibility in tracking non-standardized brain samples, albeit at the cost of offsetting some of its strengths. A customized version of NYBB tracking system has been installed at the National Prion Disease Pathology Surveillance Center at Case Western Reserve University in Cleveland, OH. This version of NYBB tracking system works with en bloc frozen samples and samples of random dimensions, by sacrificing compartment dimensions within the freezer racks, allowing for a less stringent freezer compartmentalization and tracking the estimated weight of the sample as an additional parameter. This approach was chosen to account for the relatively diminished importance of anatomical accuracy of dissection and heightened safety precautions in place when working with Prion Diseases.
Discussion
The system of standardized tissue samples (SBB; SCxV/SWmV) (Vonsattel et al. 1995, 2007) has been developed through many years of interaction of three well-established brain banks (Harvard Brain Tissue Resource Center, HBTRC, Harvard University, Boston, MA, USA; Alzheimer Disease Research Center of the Massachusetts General Hospital, Boston, MA, USA; and New York Brain Bank, NYBB, Columbia University, New York, NY, USA) and many basic science researchers in the United States and abroad. The outstanding features and advantages of this novel approach are: (1) meticulous dissection of one half of each brain donated for research at fresh state—to ensure anatomic accuracy and to harvest samples ready for immediate disbursement once categorized diagnostically; (2) perform thorough neuropathological evaluation on the formalin fixed contralateral half brain; and (3) harvesting a subset of standardized blocks from the half processed fresh and frozen, which perfectly match the subset of blocks obtained from the fixed contralateral half, which are processed for microscopic evaluation. A key component in accomplishing both is the NYBB tracking system, which allows tracking of tens of thousands of bar coded samples in NYBB freezers. Any tissue sample of the 54,242 fresh frozen tissue samples currently stored at NYBB (as of 11/2007) can be located within seconds and retrieved for disbursement immediately. Electronic confirmation of placement of samples and safeguards against data entry error, i.e. bar code labeling, ensures that none of the samples is misplaced and not retrievable.
Through providing samples, containing defined anatomical structures and well-characterized neuropathologic changes, investigators can draw from the available material in an interactive process over the WWW, often requiring little administrative support from NYBB staff. While the administrative effort per request is difficult to quantify, the increase in the number of request and disbursed samples has not altered the turnaround time of requests or necessitated additional staff at NYBB. Few custom procedures are done at NYBB; likely reflecting the comprehensiveness of the standardized sampling, thus limiting time consuming, labor intensive and cumbersome reprocessing of frozen tissue. NYBB employs one tissue-processing technician, who handles all tissue requests and manages all storage operations. A brain bank of comparable size typically employs more technicians (personal observation of senior author), mainly for carving out tissue from en bloc frozen samples.
Apart from enhancing the anatomic accuracy of the dissection, dissecting the brain at fresh state allows for the acquisition of several levels of tissue from key regions, e.g. hippocampal formation, amygdala (Alzheimer disease); mesencephalon (Parkinson disease); striatum (Huntington disease), thus increasing the yield of tissue to be disbursed, while dissection of frozen tissue is more disruptive and frequently leads to a single sample of the region of interest which is given to a single investigator.
Moving away from storing tissue as en bloc frozen samples has also made it feasible to store tissue from one donor in several freezers, which, in the invent of tissue loss due to unrecoverable failure of a single freezer, would prevent loss of all tissue from a subset of donors, thus minimizing the impact of the failure of a freezer. The compartmentalization of the freezer space has another procedural advantage: Since the compartmentalized freezer space is a known, finite entity, it is possible to determine the free space in individual freezers and all freezers of NYBB. Therefore it is possible to tailor the harvest of future samples based on the demand and the available storage space. Furthermore, since the sample collection is standardized, it is possible to focus the acquisition of future samples on regions that are underrepresented in the freezers and cut down on regions that are overrepresented, either due to increased volume of acquisition or lack of demand, thus fostering an efficient use of the available freezer space and other resources. It is difficult to accurately estimate the cost benefits of freezer management. Savings not only apply to the costs of acquiring new freezers, maintaining and operating them, but also the conservation of dissection materials and disposables, such as liquid nitrogen, freezer bags, labels, blades, gauze pads, etc., due to targeted harvesting of samples.
As research is conducted on samples obtained from NYBB, the remaining tissue samples become increasingly better characterized in terms of histological and molecular alterations present in the donated brain. This information is gradually fed back into the database and allows for an enhanced selection of suitable tissue for further studies, using previously obtained data as a criterion for inclusion of certain brains.
While working with standardized tissue samples has several advantages, NYBB tracking system has retained the flexibility to work with non-standardized samples, albeit at the cost of relinquishing some of its strength. The approach utilized at Case Western Reserve University, i.e., allowing for a less rigid freezer compartmentalization, made it possible to track samples of random dimensions. This is a useful feature when working with already processed samples that do not conform to more modern processing methods. Accommodating these samples makes usage of freezer space less efficient and necessitates handling of frozen tissue, requiring staff and prolonging the turnaround time of tissue requests; however, retrieval of tissue and keeping an accurate inventory is still accomplished with relative ease.
In summary, the NYBB system of brain banking, which has the dissection at fresh state and computer aided, bar code driven tissue tracking as its salient innovative features, has the following key advantages:
Dissection of the fresh brain allows for a more anatomically accurate dissection, thus vastly enhancing the tissue quality provided for basic science research.
Thaw-refreeze cycles with cumbersome, inaccurate and dangerous carving out of regions of interest has become obsolete, thus increasing cost effectiveness and fast turnaround time for tissue requests.
Freezer management is enhanced by compartmentalization and tissue tracking, reducing the tissue retrieval time and temperature fluctuations, which are harmful to equipment and tissue samples.
Reutilization of vacant freezer compartments with newly harvested tissue samples ensures economical use of freezer space and cost effectiveness.
Accurate quantitative data on stored samples according to anatomical regions and distributive diagnosis guides future sample collection and fosters effective use of limited resources.
Acknowledgements
This work was supported by grants from the National Institutes of Health and National Institute on Aging: P01-AG07232, R37-AG15473 and P50-AG08702; the Hereditary Disease Foundation; the Iseman Fund; and Rudin Fund.The authors would like to thank Ms. Mkeba Cason for her administrative support. The New York Brain Bank (NYBB) is especially thankful to the numerous pathologists who referred case material and to the families of the patients for providing brain tissue for research.
References
- Alafuzoff I, Winblad B. How to run a brain bank: potentials and pitfalls in the use of human post-mortem brain material in research. J Neural Transm. 1993;39(Suppl):235–243. [PubMed] [Google Scholar]
- Grinberg LT, Ferretti RE, Farfel JM, et al. Brain bank of the Brazilian aging brain study group—a milestone reached and more than 1, 600 collected brains. Cell Tissue Bank. 2007;8(2):151–162. doi: 10.1007/s10561-006-9022-z. [DOI] [PubMed] [Google Scholar]
- Ravid R, Van Zwieten EJ, Swaab DF. Brain banking and the human hypothalamus—factors to match for, pitfalls and potentials. Prog Brain Res. 1992;93:83–95. doi: 10.1016/s0079-6123(08)64565-3. [DOI] [PubMed] [Google Scholar]
- Ravid R, Swaab DF, Van Zwieten EJ, et al. Controls are what makes a brain bank go round. In: Cruz-Sánchez FF, Cuzner ML, Ravid R, editors. Neuropathological diagnostic criteria for brain banking, biomedical and health research. vol 10. IOS Press; Amsterdam: 1995. pp. 4–13. [Google Scholar]
- Schmitt A, Bauer M, Heinsen H, et al. How a neuro-psychiatric brain bank should be run: a consensus paper of Brainnet Europe II. J Neural Transm. 2007;114(5):527–537. doi: 10.1007/s00702-006-0601-8. [DOI] [PubMed] [Google Scholar]
- Stopa EG, Bird ED. Brain donation. N Engl J Med. 1989;320(1):62–63. doi: 10.1056/nejm198901053200118. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Aizawa H, Ge P, et al. An improved approach to prepare human brains for research. J Neuropathol Exp Neurol. 1995;54(1):42–56. doi: 10.1097/00005072-199501000-00006. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Del Amaya MP, Keller CE. Twenty-first century brain banking. Processing brains for research: the Columbia University methods. Acta Neuropathol (Berl) 2007;115(5):509–532. doi: 10.1007/s00401-007-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]