Figure 2.
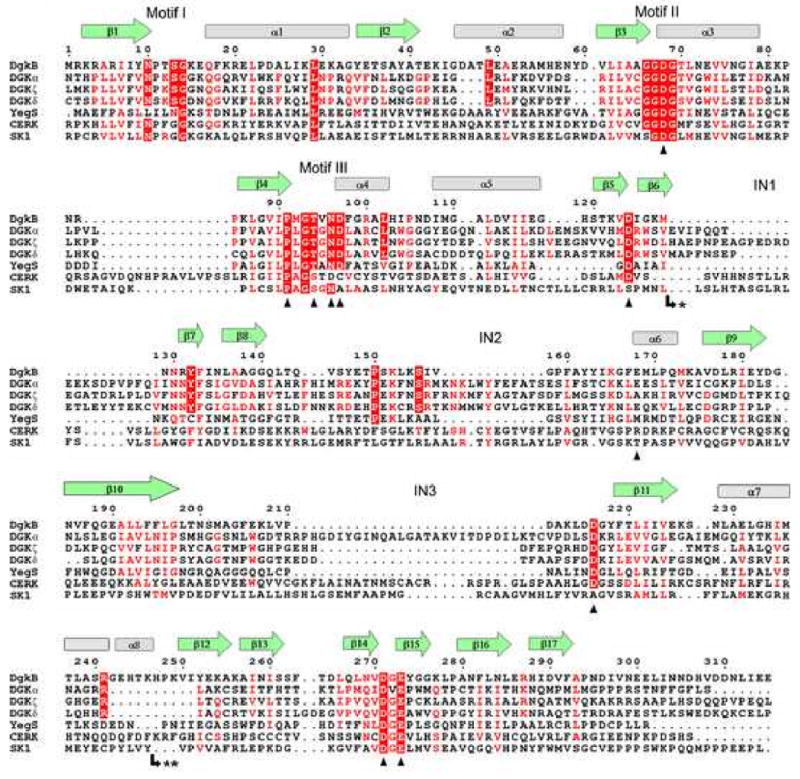
Structure-aided sequence alignment of S. aureus DgkB and human DAG kinases. The ten known isoforms of human DAG kinases were included in the similarity calculation, but only the structurally diverse DGKα, DGKζ, and DGKδ (types 1, 4, and 2, respectively) are shown for brevity. Also shown are the sequences of the related superfamily proteins E. coli YegS, human ceramide kinase (CERK) and human sphingosine kinase (SK1). DgkB β-strands and α-helices are shown as green arrows and grey cylinders, respectively. The predicted locations of the insertions in human DAG kinases are indicated by IN1, IN2, and IN3. Motifs I, II, and III refer to important ATP binding regions. Mutational analyses were performed on DgkB residues marked with a black triangle. The most highly conserved residues are in red block, and less conserved residues are indicated by red font. Note: * marks the insertion of an extra domain in type 2 DGKs (DGKδ, DGKη, and DGKκ comprising 296, 286, and 214 amino acids, respectively) and ** marks the insertion of 30 amino acids in CERK.
