Figure 10.
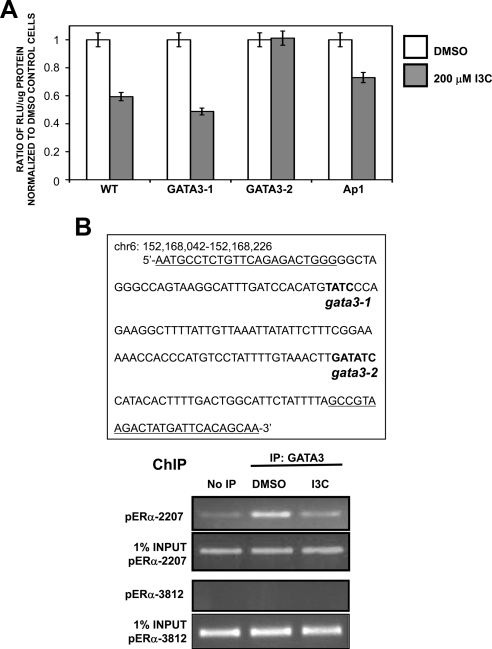
Identification of GATA3 as the transcription factor responsible for ERα promoter down-regulation by I3C. (A) MCF-7 cells were transfected with luciferase reporter plasmids driven by either the wild-type ERα promoter fragment starting at −2294 upstream of the RNA site start, or with ERα promoter fragments with the designated mutations in the consensus GATA3-1, GATA3-2, or Ap1 transcription factor binding sites. Twenty-four hour-transfected cells were treated for an additional 24 h with either DMSO or 200 μM I3C. Relative luciferase activity was evaluated in lysed cells using the Luciferase assay kit (Promega). Bar graphs indicate relative luciferase activity normalized to the protein input. Values are normalized to DMSO of each transfection. Result was repeated twice. (B) Left, ERα genomic sequence containing both predicted GATA3 binding sites (bold) within the I3C-responsive region of ERα promoter. Primers used to amplify GATA3 sites for chromatin immunoprecipitation are underlined. Chromatin was isolated from MCF-7 cells treated with or without 200 μM I3C for 24 h. GATA3 was immunoprecipitated from total cell extracts using Sepharose G bound to anti-ERα antibody. DNA released from ERα was amplified using indicated primers. Control primers directed at upstream site (−3812 base pairs) showed no amplification in IP samples. Input samples represent total genomic DNA from each treatment (loading control). This result was repeated twice.