Abstract
Infrared multiphoton dissociation (IRMPD) on a linear ion trap mass spectrometer is applied for the sequencing of small interfering RNA (siRNA). Both single-strand siRNAs and duplex siRNA were characterized by IRMPD, and the results were compared to that obtained by traditional ion trap based collision induced dissociation (CID). The single-strand siRNA anions were observed to dissociate via cleavage of the 5’ P–O bonds yielding c- and y-type product ions as well as undergo neutral base loss. Full sequence coverage of the siRNA anions was obtained by both IRMPD and CID. While the CID mass spectra were dominated by base loss ions, accounting for ~25 – 40% of the product ion current, these ions were eliminated through secondary dissociation by increasing the irradiation time in the IRMPD mass spectra to produce higher abundances of informative sequence ions. With longer irradiation times, however, internal ions corresponding to cleavage of two 5’ P–O bonds began to populate the product ion mass spectra as well as higher abundances of [a - Base] and w-type ions. IRMPD of siRNA cations predominantly produced c- and y-type ions with minimal contributions of [a - Base] and w-type ions to the product ion current; the presence of only two complementary series of product ions in the IRMPD mass spectra simplified spectral interpretation. In addition, IRMPD produced high abundances of protonated nucleobases – [G+H]+, [A+H]+, and [C+H]+ - which were not detected in the CID mass spectra due to the low-mass cut-off associated with conventional CID in ion traps. CID and IRMPD using short irradiation times of duplex siRNA resulted in strand separation, similar to the dissociation trends observed for duplex DNA. With longer irradiation times, however, the individual single-strands underwent secondary dissociation to yield informative sequence ions not obtained by CID.
INTRODUCTION
The identification and determination of the function of small non-coding (i.e., non-messenger) RNA including small interfering RNA (siRNA) and micro-RNA (miRNA) has become an important challenge due to the growing recognition of these RNAs in fundamental cellular processes including gene regulation, RNA processing, translocation of proteins, chromosome maintenance, and cancer and disease development [1, 2]. For example, RNA interference (RNAi) is a biological pathway which down-regulates or silences gene expression and has shown great potential to be harnessed as a therapeutic tool via introduction of siRNA, typically short double stranded RNA consisting of approximately 25 nucleotides (per single strand), to cells [3, 4]. Micro-RNAs, which consist of ~22 nucleotides, have also been shown to effect gene silencing by binding directly to the target mRNA [5]. Several siRNA-based drugs are currently in clinical trials [6, 7], and recent studies have explored the impact of siRNA on coronary artery disease [8], Alzheimer’s disease [9] , and HIV [10, 11].
The traditional methods used to identify and sequence oligonucleotides include cloning and expression techniques such as the Sanger method, base-specific chemical cleavage methods, reverse transcription, and RNA fingerprinting for identifying modifications [12, 13]. More recently, deep sequencing analyses in conjunction with predictive computational approaches have also been utilized to identify miRNAs [14, 15]. Compared to these traditional gel electrophoresis-based techniques, mass spectrometry provides accurate mass determinations as opposed to relative migration values and affords more rapid analysis [12, 16]. Limbach et al. have used mass spectrometry along with endonucleases to identify RNAs based on their unique digestion products [17–20].
Tandem mass spectrometry has been widely used to sequence DNA [21–26], and in recent years more efforts have been devoted to the analysis of RNA [27–32]. Gas-phase dissociation of DNA anions is initiated by base loss and subsequent dissociation of the 3’ C–O bond yielding complementary [a - Base] and w-type ions [22]. In contrast, collision-induced dissociation (CID) of RNA typically results in fragmentation of the 5’ P–O bond producing complementary c- and y-type fragment ions [27–30]. Schürch and co-workers observed that dissociation of RNA was independent of the nucleobase and that cleavage of the 5’ P–O bond is related to the presence of the 2’-hydroxyl group [28, 30]. That is, upon ion activation, an intramolecular cyclic transition-state involving the 2’-hydroxyl proton and the 5’ oxygen of the phosphate is formed, and migration of the 2’-hydroxyl proton and subsequent cleavage yields the c- and y-type fragments [28, 30]. McLuckey’s group observed that the dissociation pathways of RNA anions were charge-dependent in which more highly charged species predominantly underwent base loss, and the lower charge states gave higher sequence coverage [27]. More recently, McLuckey et al. obtained full sequence coverage of deprotonated siRNA ions by CID with subsequent proton transfer reactions which effectively “deconvoluted” and simplified interpretation of the product ion mass spectra [31]. The Håkansson group investigated the fragmentation pathways of oligoribonucleotides by both electron capture dissociation (ECD) and electron detachment dissociation (EDD) [33]. They observed that the ECD fragmentation patterns were dependent on the nucleobase. Both c/y-type ions and d/w-type ions were produced by ECD; whereas EDD predominantly yielded d- and w-type ions, and full sequence coverage was obtained.
Tandem mass spectrometry is likewise well-suited for the characterization of novel miRNAs, as well as synthesized siRNAs, including those that contain chemical modifications to increase their stability, such as deoxynucleotides, 3’-overhangs, or 2’-O-methyl modifications [34, 35]. Herdewijn and co-workers identified post-translational modifications of ribosomal RNA by mass spectrometry [36, 37]. The Fabris lab has combined chemical probes in conjunction with tandem mass spectrometric analyses to probe RNA structures [38–41]. Recently, Karas’s group degraded RNA by acid hydrolysis of the phosphodiester bonds to produce 5’- and 4’-end mass ladders for sequencing of 21-mers by matrix-assisted laser desorption ionization mass spectrometry [42].
Infrared multiphoton dissociation (IRMPD) has been previously been shown to be a viable alternative and complementary dissociation method to CID for the sequencing of DNA [23, 24, 43–45]. One of the primary advantages of IRMPD in ion traps is that as a non-resonant method, the precursor ion and all ensuing product ions are activated by IR irradiation. Uninformative product ions due to dead-end dissociation pathways (e.g., neutral base loss) can undergo secondary dissociation to yield diagnostic sequence ions. IRMPD also does not suffer significantly from the low-mass cut-off problem intrinsic to conventional CID in rf ion traps. The energy deposition by conventional CID is dependent on the rf trapping voltage which prevents the storage of ions of m/z in the lower 28% of the mass range relative to the precursor ion. Since IRMPD can be performed using far lower rf trapping levels, diagnostic low-mass product ions can be detected. The energy deposition by photon absorption is also not dependent on the rf trapping voltage, but rather on the absorptivity of the ion at the wavelength of the laser. Compared to proteins and peptides which do not undergo efficient photodissociation at normal trap pressures of ~ 1 mTorr, oligonucleotides dissociate rapidly upon exposure to IR irradiation at 10.6 µm due to the high IR absorptivity of the phosphodiester backbone at this wavelength [46], an effect noted initially by Little et al. [47]. IRMPD also affords a high degree of tunability and control with regards to energy deposition, as the irradiation time and power can both be varied.
In the present study, we examine the use of IRMPD to sequence single-strand and duplex siRNA anions and cations as a function of charge state. Time-resolved IRMPD experiments reveal the genealogy of product ion formation. Compared to CID, IRMPD produces a higher abundance of diagnostic sequence ions and favors the formation of shorter product ions in lower charge states. Full sequence coverage was generally obtained by both dissociation techniques; however, the IRMPD product ion mass spectra were often simpler to interpret due to conversion of the neutral base loss product ions into informative sequence ions upon secondary dissociation.
EXPERIMENTAL
Chemicals and Materials
Imidazole, piperidine, and Amberlite IRN77 resin were purchased from Sigma-Aldrich (St. Louis, MO). Methanol, ammonium acetate, and isopropanol were obtained from Fisher Scientific (Fairlawn, NJ). The siRNAs targeting the glyceraldehydes 3-phosphate dehydrogenase (GAPDH) gene and the Lamin A/C gene were purchased from Integrated DNA Technologies (Coralville, IA). The sequences for anti-GAPDH were 5’OH-GUGGAUAUUGUUGCCAUCAdTdT-3’OH (sense strand, GAPDH.S, 6633.9 Da) and 5’OH-UGAUGGCAACAAUAUCCACdTdT-3’OH (antisense strand, GAPDH.AS, 6644.9 Da). The sequences for anti-Lamin A/C were 5’-biotin-GGAACUGGACUUCCAGAAGAACAUCUA-3’OH (sense strand, Lamin A/C.S, 9053.6 Da) and 5’-UAGAUGUUCUUCUGGAAGUCCAGUUCC (antisense strand, Lamin A/C.AS, 8544.1 Da).
Single-strand siRNA samples were desalted by cation exchange using the Amberlite IRN77 resin as described by McLuckey et al. [31]. Duplex siRNA was annealed by combining the sense and antisense siRNA strands (30 µM each) in 150 mM NH4OAc, pH 7.0. The samples were heated at 90 °C for three minutes and then cooled to room temperature overnight. Duplex siRNA solutions were diluted to 10 µM in 25:75 (v/v) methanol/50 mM NH4OAc in nano-pure H2O. Solutions of GAPDH single-strand siRNAs were prepared at 10 µM in either 25:75 (v/v) methanol/25 mM NH4OAc for negative and positive electrospray mass spectrometry (ESI-MS) or 20:80 (v/v) isopropanol/H2O with 25 mM imidazole and 25 mM piperidine for negative ESI-MS. The Lamin A/C single strand siRNAs were prepared at 10 µM in 25:75 (v/v) methanol/25 mM NH4OAc for negative and positive ion ESI-MS.
Mass Spectrometry and Infrared Multiphoton Dissociation
All experiments were conducted on a modified LTQ XL linear ion trap mass spectrometry (Thermo Fisher Scientific, San Jose, CA) with the standard electrospray ionization (ESI) source. Samples were infused at 2.5 µL/min and the ESI voltage was set to either −3.5 or +4.0 kV for negative and positive ion mode analysis, respectively. The inlet heated capillary was maintained at 90 °C. The mass spectrometer was modified to perform infrared multiphoton dissociation as previously described [48]. Briefly, a ZnSe window was mounted to the back plate of the vacuum manifold to transmit 10.6 µm radiation from a model 48-5 Synrad 50 W cw CO2 laser (Mukilteo, WA). The laser was triggered through the LTQTune v.2.5.0 software during the activation step in the scan function. The irradiation time was varied between 0.0 and 20.0 ms and the power was maintained at 10 W (25%). The q-value for all IRMPD experiments was set between 0.055 and 0.100. Collision-induced dissociation was performed at the typical q-value of 0.25, the activation time was set to 30 ms, and the normalized collision energy was adjusted to dissociate the precursor ion to less than 1% relative abundance.
Data Analysis
Time-resolved IRMPD experiments were performed by incrementally increasing the irradiation time and the abundances of the precursor and product ions were plotted against the irradiation time. Ion abundances were determined by measuring the peak areas using Origin 7.0. Product ions were identified using Mongo Oligo Mass Calculator (http://library.med.utah.edu/masspec/mongo.htm) in addition to an oligonucleotide product ion calculator program written in-house using National Instruments LabView 8.5. Only product ions which could be unambiguously identified were included in the product ion type abundance calculations. Product ions were labeled according to the nomenclature proposed by McLuckey et al. [22].
RESULTS AND DISCUSSION
Infrared Multiphoton Dissociation vs. Collision-Induced Dissociation of Single-Strand siRNA
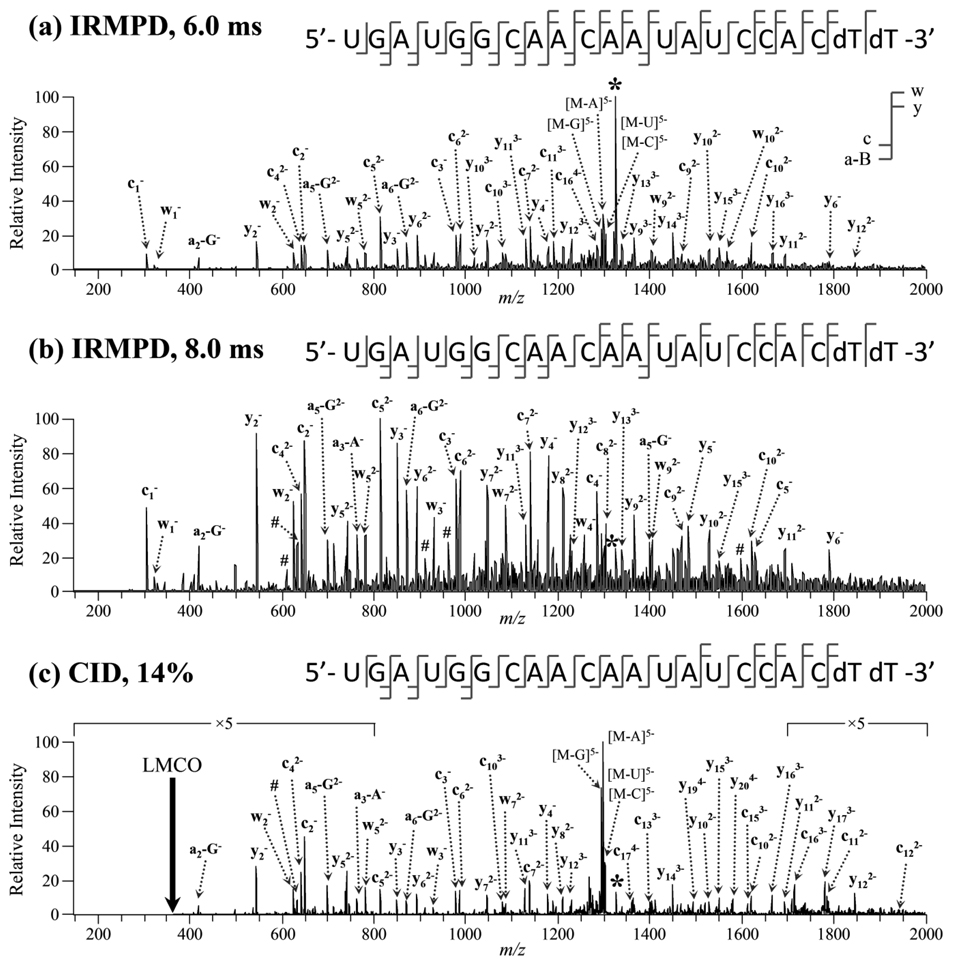
In the negative ion mode, ESI of the GAPDH sequences in ammonium acetate buffer produced ions in the 4− to 6− charge states, while ESI of Lamin A/C samples in ammonium acetate yielded ions predominantly in the 5− and 6− charge states (data not shown). Electrospray ionization of the GAPDH sequences in the imidazole/piperidine additive solution produced ions in charge states ranging from 6− to 10−. In the positive mode, both the GAPDH and Lamin A/C sequences produced ions in only two charge states. The single-strand siRNAs were subjected to IRMPD, and the product ion mass spectra were compared to that obtained by conventional ion-trap CID. Examples of the IRMPD and CID mass spectra are shown in Figure 1 and Figure 2. Sannes-Lowery and Hofstadler have previously analyzed an oligoribonucleotide by IRMPD in an external hexapole by FT-ICR-MS [43], but IRMPD has not been extensively investigated as an alternative dissociation method for RNA. The fragmentation patterns of siRNA anions by IRMPD is dependent on the irradiation time (i.e., photon flux), and thus by varying the time (or laser power), the product ion mass spectra can be effectively tuned by controlling the extent of secondary dissociation. IRMPD of the 5− charge state of GAPDH.S using 6.0 ms irradiation predominantly yielded c- and y-type product ions due to cleavage of the 5’ P–O bond (Figure 1a). Lower abundance [a - Base] and complementary w-type ions were also detected. The abundances of product ions stemming from neutral base loss, which typically dominate CID product ion mass spectra of RNA [27], were modest. Almost a complete series of c-type (19/20) and y-type (18/20) ions was observed for this siRNA anion. The three c and y product ions which were not observed were y1, its complementary c20 ion, and y20. Cleavage between the two dT nucleotides would yield the y1 and c20 ions, and these product ions are not likely to be observed due to the lack of the 2’-OH group to facilitate cleavage of the 5’ P–O bond. However, the w1 ion, which corresponds to cleavage of the 3’ C–O bond, was detected, and full sequence coverage of this siRNA anion was obtained. The complementary [a20 - T] and the y20 product ions were most likely not detected due to secondary dissociation of these large product ions. Upon increasing the irradiation time to 8.0 ms, the product ion mass spectra changes significantly (Figure 1b). The more highly-charged and larger product ions observed in Figure 1a undergo secondary dissociation into smaller product ions. Both the precursor ion and base loss product ions are almost completely dissociated, and the product ion mass spectrum is dominated by c- and y-type fragment ions. A majority of the [a - Base] ions detected are due to the facile loss of neutral guanine (i.e., [an - G] product ions predominant) which has previously been reported to be a significant dissociation pathway for RNA anions in low charge states [27]. The [a - Base] and w-type product ions are likely produced by secondary photodissociation of the neutral base loss product ions (e.g., [M - G]5−) and subsequent cleavage of the 3’ C–O bond of the ribose from which the base was lost. Full sequence coverage of GAPDH.S was obtained using 8.0 ms of irradiation even though fewer sequence ions were detected (c1 – c12 and y2 – y15). The lack of larger, more highly-charged product ions reduces the spectral complexity while still allowing the siRNA anion to be completely sequenced. A majority of the product ions were singly or doubly charged. Several low mass, singly charged product ions which could be not be identified as sequence (i.e., [a - Base], c-, y-, w-type) ions were also detected. These product ions were determined to be internal ions due to cleavage of two 5’ P–O bonds. These internal ions could not be unambiguously identified due to isobaric species (e.g., AUG = GAU). The abundances of the internal ions are low, and the ions likely arise from secondary dissociation of larger c- and y-type product ions. The CID product ion mass spectrum of [GAPDH.S – 5H]5− was dominated by neutral base loss (Figure 1c). Product ions were observed in all possible charge states and almost complete sequence coverage was obtained. Due to the low-mass cut-off (LMCO) problem inherent to conventional CID, the low mass c1 (m/z 305) and w1 (m/z 324) sequence ions could not be detected. Fewer [a - Base] and w-type ions were observed upon CID, a result attributed to the fact that only the precursor ion is activated and primary product ions such as [M-G] are not further dissociated into sequence informative species. Small internal ions were also detected by CID; however, these product ions were not as abundant as those in the IRMPD mass spectrum obtained using 8.0 ms of irradiation.
Figure 1.
IRMPD product ion mass spectra of [GAPDH.S – 5H]5− of m/z 1326.0 using (a) 6.0 ms and (b) 8.0 ms of irradiation at 10 W at q-value of 0.10. (c) CID product ion mass spectra of the same siRNA anion (14% normalized collision energy, q = 0.25). The precursor ion is indicated by an asterisk (*) and internal ions by #. Not all peaks were labeled to reduce congestion.
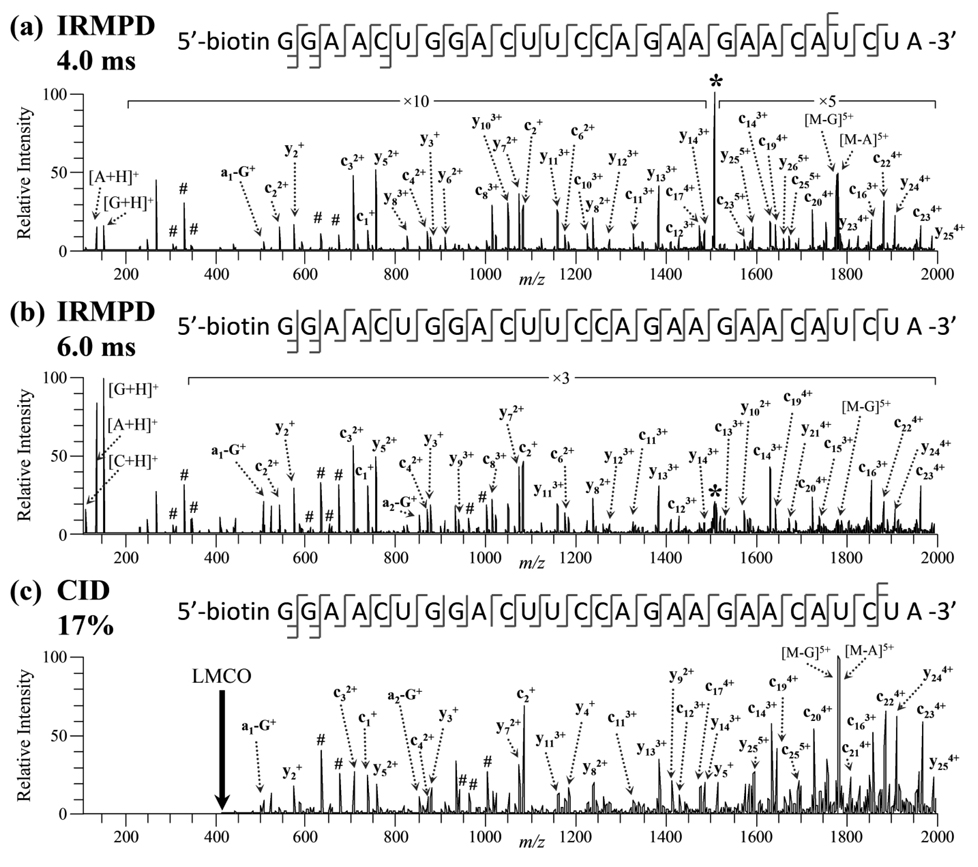
Figure 2.
IRMPD product ion mass spectra of [Lamin A/C.S + 6H]6+ of m/z 1509.8 using (a) 4.0 ms and (b) 6.0 ms of irradiation at 10 W at q-value of 0.065. (c) CID product ion mass spectra of the same siRNA cation (17% normalized collision energy, q = 0.25). The precursor ion is indicated by an asterisk (*) and internal ions by #. Not all peaks were labeled to reduce congestion.
IRMPD mass spectra of the 6+ charge state of the sense strand of Lamin A/C using 4.0 and 6.0 ms of irradiation are shown in Figures 2a and 2b. At short irradiation times, almost complete series of both c-ions (c1 – c25) and y-ions (y2 – y26) were observed. In contrast to dissociation of siRNA anions, the product ions were predominantly composed of c/y-type fragments, and the mass spectrum was almost completely void of [a - Base] and w-type ions. Also, siRNA cations of all charge states dissociated through protonated base loss, and the resulting protonated bases were observed in the low m/z range of the IRMPD mass spectrum. The [Base + H]+ ions were the most abundant product ions in the mass spectrum with protonated guanine observed at higher abundance than [A + H]+ and [C + H]+; [U + H]+ and [T + H]+ were not detected in the IRMPD mass spectra of any of the siRNA cations. The relative abundances of the protonated nucleobases are not directly proportional to the relative contribution of each nucleobase to the sequence composition. In fact, the position of the nucleobases and their relative gas-phase basicities influence the abundance of the protonated base product ions as well. Since uracil and thymine have the lowest gas-phase proton affinities of the five nucleobases [49–51], the absence of these protonated bases in the IRMPD spectra is not surprising. Low mass internal ions were also detected in the IRMPD mass spectrum. Increasing the irradiation time to 6.0 ms led to significantly higher abundances of the three protonated nucleobases, [G + H]+, [A + H]+, and [C + H]+, as shown in Figure 2b. Identical sequence coverage was obtained using 6.0 ms of irradiation as 4.0 ms of irradiation, but base loss ions (e.g., [M - G]5+ and [M - A]5+) were significantly reduced in abundance, and a higher abundance of internal ions were produced. With even longer irradiation times, sequence ions were converted almost exclusively into protonated bases (data not shown). The CID mass spectrum of [Lamin A/C.S + 6H]6+ was dominated by protonated base loss, but due to the LMCO problem, the protonated base ions were not detected (Figure 2c). While the spectrum is more cluttered than the IRMPD mass spectra, full sequence coverage was still obtained. The c/y ions were prominent, but almost no [a - Base] and w-type ions were detected upon CID of siRNA cations. Interestingly, a higher abundance of internal ions was observed upon CID of the siRNA cations than the anions.
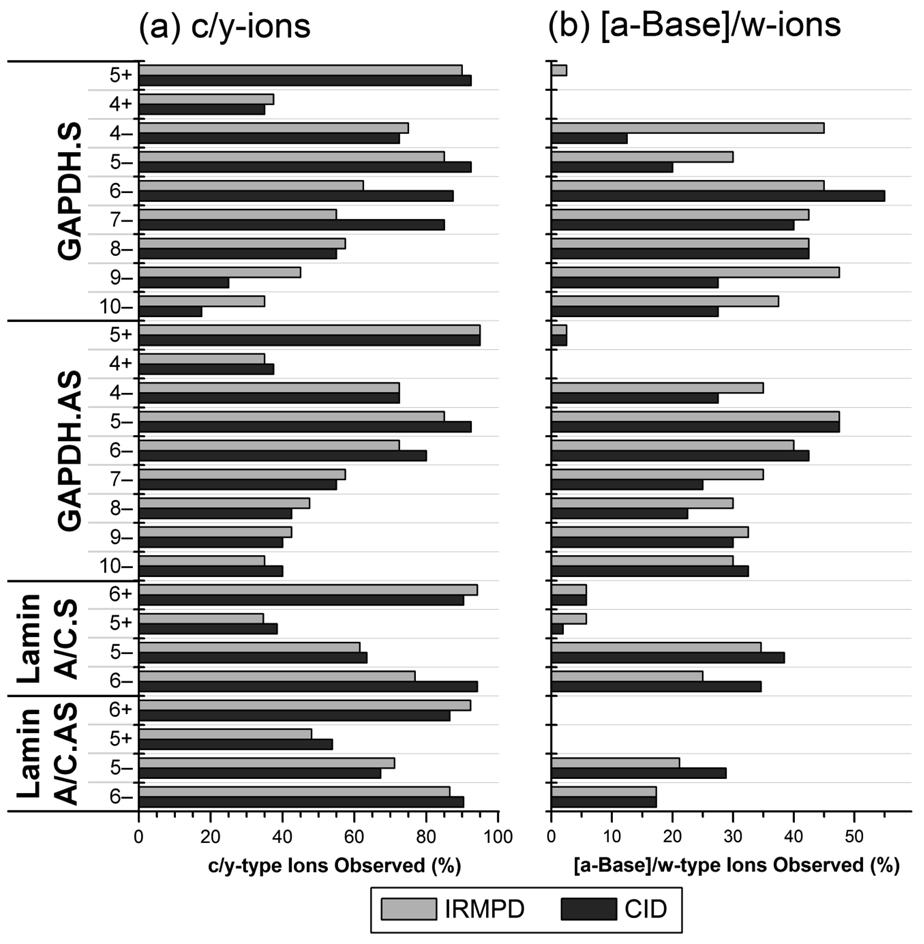
The percentages of c/y-type ions and [a - Base]/w-type ions observed by IRMPD and CID arising from IRMPD and CID are summarized for all of the siRNA ions in Figure 3. In general, both IRMPD and CID yielded similar degrees of sequence coverage based on the number of unique product ions. As the “charge level” (defined as the number of negative charges per phosphate for oligonucleotide anions [52] or the number of positive charges per nucleobase for cations) of the anions increased above 33% (i.e., 7− through 10− charge states of GAPDH.S and GAPDH.AS), the percent of c/y-type ions observed by IRMPD and CID decreased as many product ions could not be unambiguously identified due to isobaric overlaps and the inability to isotopically resolve the peaks to determine the charge state. For charge levels of the siRNA anions below 33% (4− through 6− charge states), near complete series of c/y-ions were detected, and CID generally outperformed IRMPD as the longer c- and y-type ions were often not detected in the IRMPD mass spectra likely due to rapid secondary dissociation. The percent of [a - Base]/w-ions was generally between 20 and 50% for the siRNA anions studied, and a higher number of these product ions were detected in the IRMPD mass spectra. This trend is attributed to secondary dissociation of base loss product ions by IRMPD as discussed earlier.
Figure 3.
(a) Percent of c- and y-type ions and (b) percent of [a - Base] and w-type ions of model single-strand siRNAs in various charge states observed by IRMPD (light gray) and CID (dark gray). The data was compiled from IRMPD mass spectra in which the precursor ion was reduced to ~10% relative abundance (required 5 – 7 ms of irradiation). A value of 100% would indicate that all possible c/y-ions or [a - Base]/w-ions were detected.
In the positive ion mode, only two charge states of each the single-strand siRNAs were observed in the ESI mass spectra. For the siRNA cations in all charge states examined, less than 10% of the possible [a - Base]/w-type ions were detected by both IRMPD and CID (Figure 3). At charge levels above 20% (5+ charge state of GAPDH and 6+ charge state of Lamin A/C siRNAs), near complete series of c/y-ions were detected by both dissociation methods, but at charge levels below 20% (4+ charge state of GAPDH and 5+ charge state of Lamin A/C siRNAs) less than 40% of the possible c/y-ions were observed. IRMPD mass spectra of [Lamin A/C.AS + 5H]5+ and [GAPDH.AS + 4H]4+ are shown in Supplemental Figure 1a and 1b. Fewer than 30 and 20 product ions were identified in the two product ion mass spectra, respectively, and only the 5’ and 3’ ends of the siRNAs were sequenced. Between six and ten nucleotides from either terminal were sequenced for the GAPDH and Lamin A/C siRNAs. For all of the siRNA cations in the lowest charge state (5+ for the Lamin A/C sequences and 4+ for the GAPDH sequences), no product ions stemming from cleavage of the phosphate backbone near the middle of the oligonucleotide were detected (typically the middle ten nucleotides). A summary of the product ions observed by CID and IRMPD of these siRNA cations in low charge states is shown in Scheme 1. Almost identical results were obtained by CID as well (data not shown). The low sequence coverage of these siRNAs may be due in part to the high m/z value of the precursor ion. Upon cleavage of the phosphate backbone, some of the expected product ions would have m/z values greater than 2000 due to their low charge states. The lack of product ions arising from dissociation near the center of the siRNAs does not seem to be nucleobase dependent as this effect was observed for all four of the siRNAs which each have a different composition of nucleobases in this region. These results may also be due to the slow-heating methods (i.e., IRMPD and CID) employed on the linear ion trap mass spectrometer and greater sequence coverage of these siRNA cations may be obtained on other dissociation techniques (e.g., ETD, surface-induced dissociation, beam-type CID).
Scheme 1.
Observed bond cleavages of siRNA cations in the lowest observed charge states.
Product Ion Abundances and Time-resolved IRMPD of Single-Strand siRNA
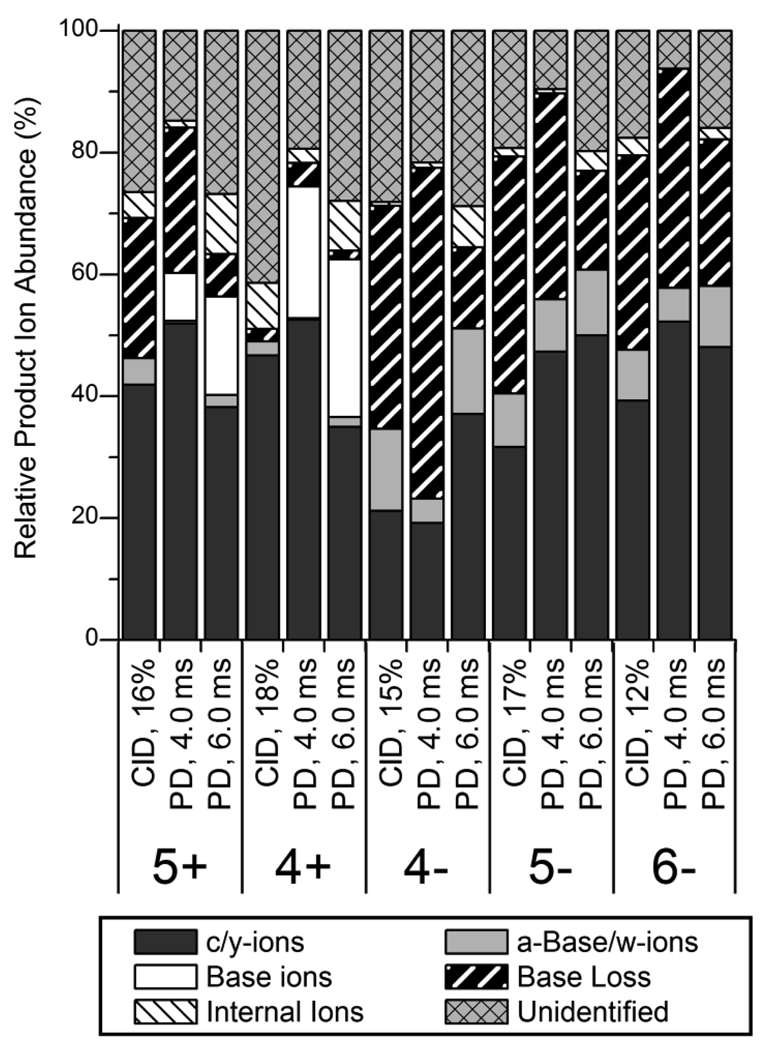
The contributions of each type of product ion to the total product ion current in IRMPD and CID mass spectra of GAPDH.AS in the 6−, 5−, 4−, 4+ and 5+ charge states were calculated and summarized in Figure 4. (The more negatively charged GAPDH.AS precursor ions – 7−, 8−, 9− and 10− charge states – were not included in this analysis because many ions could not be unambiguously identified as the isotopic profiles of the more highly charged product ions could not be resolved.) In general, approximately 40 – 50% of the product ion current was composed of c- and y-type ions; the lone exception was the 4− charge state of GAPDH.AS which was dominated by neutral base loss. For the GAPDH.AS anions, [a - Base] and w-type ions comprised ~5 – 10% of the product ion current. It should be noted that a majority of the [a - Base] and w-type ions of GAPDH siRNAs and Lamin A/C siRNAs contained fewer than 9 or 12 nucleotides, respectively, suggesting that these fragments are produced by dissociation of base loss fragment ions. Using short irradiation times (4.0 ms), the distribution of product ions in the IRMPD mass spectra were almost identical to that of the CID mass spectra. Longer irradiation times yielded lower abundances of base loss ions and higher abundances of both internal ions and protonated base ions (for the GAPDH.AS cations). Base loss ions were not observed for GAPDH.AS in the 4+ charge state as these product ions (e.g., [M-G]3+, m/z 2165.6), which are due to loss of protonated bases, have m/z values greater than that of the mass range of the linear ion trap. Similar results were also observed for the Lamin A/C siRNA single strands – CID yielded more abundant base loss ions than IRMPD using irradiation times of greater than 6 ms (data not shown).
Figure 4.
Relative abundances of different product ion types of GAPDH.AS in various charge states observed by IRMPD at 4.0 and 6.0 ms of irradiation and by CID. The abundances are relative to the total product ion abundance in the product ion mass spectra.
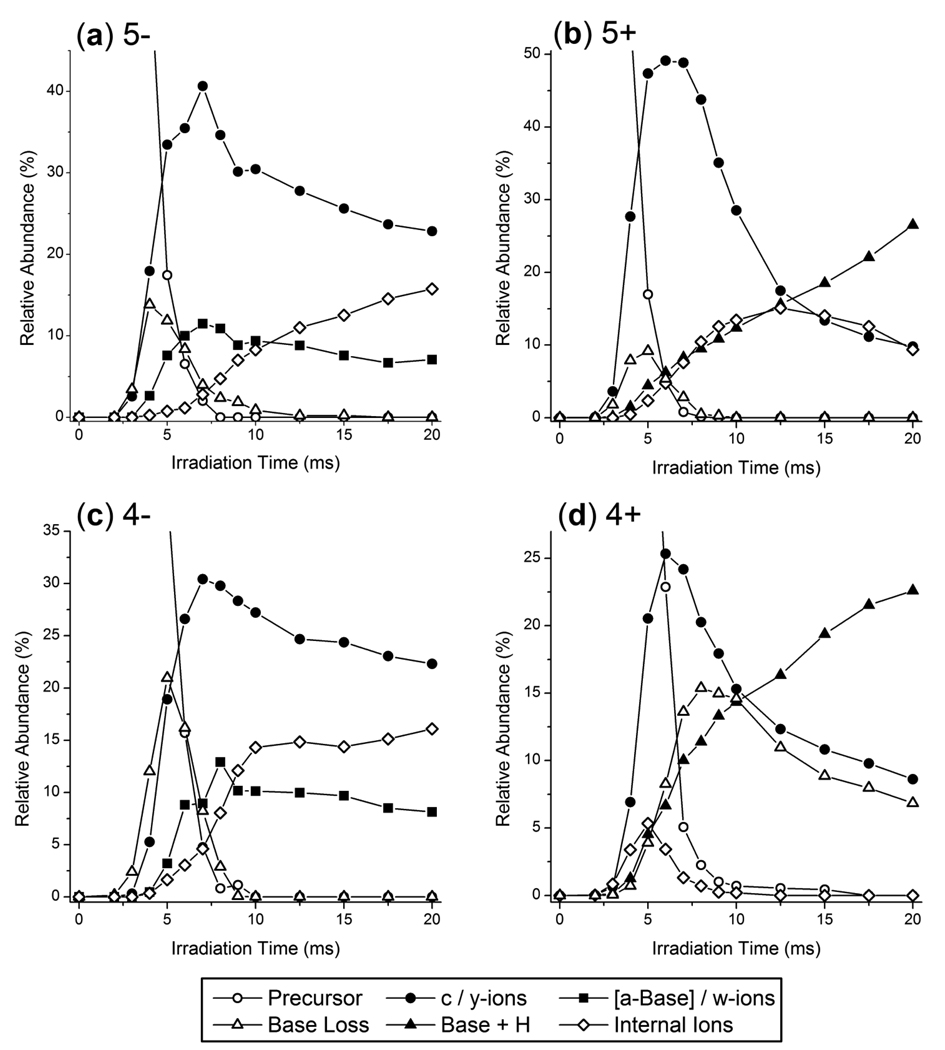
The abundances of each of these product ion types was also monitored as a function of irradiation time as shown in Figure 5 for the 5−, 5+, 4− and 4+ charge states of GAPDH.S. Base loss ions are initially observed at irradiation times of ~2 ms while c- and y-type ions begin to appear at irradiation times of ~3 ms for the GAPDH.S anions (Figure 5a and 5c). However, [a - Base] and w-type ions appear after using longer irradiation times, which provides evidence that a portion of these products ions may be secondary fragment ions of base loss species or larger c/y-type ions. The neutral base loss anions [GAPDH.S – G – 5H]5− and [GAPDH.S – A – 5H]5− were directly interrogated by CID and a wide variety of product ions were observed including expected [a-Base]/w-ions as well as y-Base, y-, and c-type ions (data not shown). These results suggest [a-Base]/w-ions are observed as secondary product ions but are likely also produced directly from the intact siRNA anion precursor. As the irradiation time increased, a maximum relative abundance of base loss ions was observed at ~5 ms at which point the abundance of base loss ions decreased at a similar rate to that of the precursor ion. After 8 ms of IR irradiation, the precursor ion was completely dissociated and base loss ions accounted for less than 3% of the total ion current in the IRMPD mass spectra. Since the precursor ion and the base loss ions contain the same number of chromogenic phosphate groups and, for the GAPDH.S anions, the same charge, it is not unexpected that both of these species would photodissociate at a similar rate. For the GAPDH.S siRNA anions, c- and y-type ions accounted for more than 30 – 40% of the total ion current at irradiation times of ~7 ms at which point their abundances decreased due to secondary dissociation. The complementary [a - Base] and w-type ions comprised between 10 and 15% of the total ion current at their maximum abundance for the siRNA anions. In contrast, c/y-type ions accounted for upwards of 50% of the total ion current in the IRMPD mass spectra of [GAPDH.S + 5H]5+ and only 25% for [GAPDH.S + 4H]4+ (Figure 5b and 5d). Base loss ions and c/y-type ions for the GAPDH.S cations were initially observed at ~3 ms of irradiation whereas the onset for [Base + H]+ ions was at ~4 ms. The abundance of protonated bases increased almost linearly with irradiation time for the GAPDH.S siRNA cations. Internal ions were also observed to increase in abundance with time, but at long irradiation times their abundance tapered off for the 5+ charge state of GAPDH.S as these ions were likely further dissociated into protonated base product ions.
Figure 6.
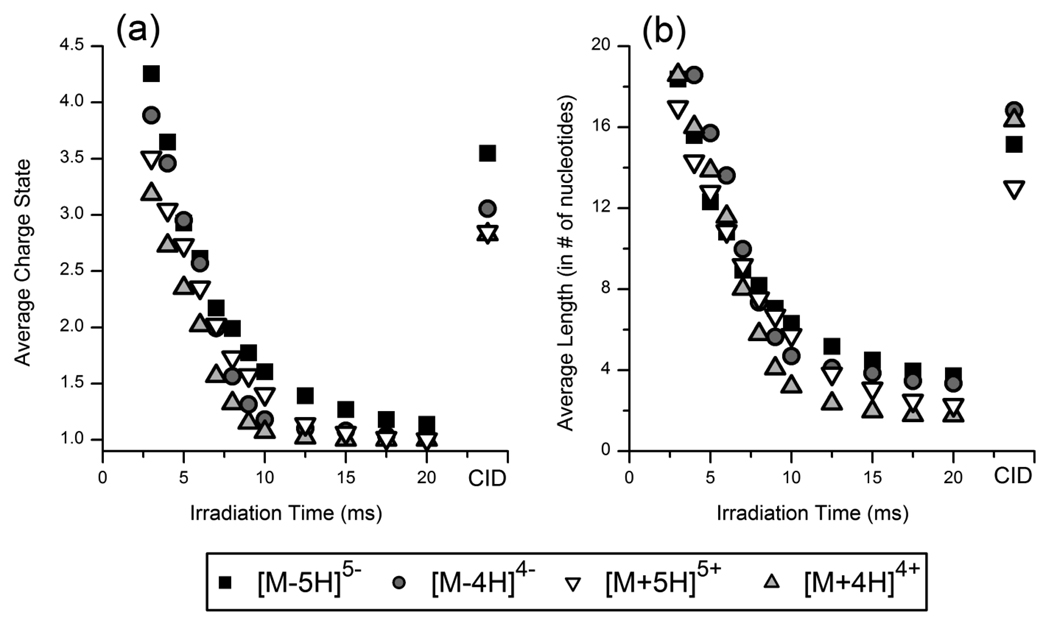
(a) Average charge state (absolute value) and (b) average length (in # of nucleotides) of product ions of GAPDH.S in the 5− (squares), 5+ (upside-down triangles), 4− (circles) and 4+ (triangles) charge states as a function of irradiation time. Values from the CID mass spectra are also plotted as the right most points in each panel.
Figure 5.
Relative abundances of product ions and the precursor ion as a function of irradiation time for the (a) 5−, (b) 5+, (c) 4−, and (d) 4+ charge states of GAPDH.S. Abundances are relative to the total ion current in each IRMPD product ion mass spectrum.
The average charge state and the average length (in number of nucleotides) of the product ions of GAPDH.S were also plotted as a function of irradiation time for the IRMPD mass spectra, in addition to the values obtained from the CID mass spectra in Figure 6. The complete charge state distribution plots and relative abundances of product ions of varying nucleotide length are shown in Supplemental Figures 2 and 3 for the GAPDH.S anions and cations, respectively. Both the average charge state and the average product ion length decreased exponentially as the irradiation time increased. As the abundance of more highly-charged product ions decreased, the abundance of product ions in lower charge states correspondingly increased due to secondary dissociation of the more highly-charged species. At irradiation times less than 4 ms, a majority of product ions of [GAPDH.S – 5H]5− were in the 5−, 4− and 3− charge states, but at 7 ms, doubly charged product ions dominated the IRMPD mass spectra. Singly charged product ions were predominantly observed at irradiation times of greater than 10 ms, with negligible contributions from ions in the 5−, 4−, and 3− charge states were negligible. In fact, greater than 90% of the identified product ions were singly or doubly charged after 10 ms irradiation. At 20 ms of irradiation time, 89% of the identified product ions were singly charged. The same trends were also observed for the Lamin A/C siRNAs – long irradiation times yielded product ions in lower charge states and with fewer nucleotides (data not shown).
While these trends suggest that more highly charged product ions undergo more rapid secondary dissociation potentially due to a greater charge mobility, other factors are likely in play. In particular, one must note that product ions observed in higher charge states also are generally greater in size (i.e., number of nucleotides) and have a greater number of phosphate chromophores. Upon examination of the relative abundances of products ions of varying length vs. irradiation time, a similar trend is observed (Figure 6b). Product ions containing all 21 nucleotides (i.e., base loss ions) were initially observed as the most abundant fragment ion at irradiation times less than 5 ms. As the irradiation increased, the abundance of longer product ions decreased and product ions containing fewer nucleotides increased. As the irradiation time increased from 7 to 9 ms, the average length of the product ions of the 5− charge state of GAPDH.S was reduced from ~9 to ~7 nucleotides but at 20 ms of irradiation, less than 8% of the total ion current was composed of product ions of greater than 6 nucleotides. For the GAPDH.S cations, the abundance of product ions with fewer than 6 nucleotides actually decreased at irradiation times greater than 10 – 12 ms as these small product ions were dissociated into protonated base ions. These trends suggest that both the charge state and the number of chromophores (i.e., phosphate groups) influence the rate of IR photodissociation of oligoribonucleotides.
The IRMPD results using a short and a longer irradiation time complement each other in that each provide different levels of sequence confirmation. Short irradiation times predominantly yield large product ions in high charge states and a minimal abundance of [a - Base] and w-type ions which simplifies spectral interpretation as one only has to search for a single ion type. By using longer irradiation times, one can more easily unambiguously identify the product ions as a majority of the species will be singly or doubly charged. Also, the presence of more [a – Base] and w-type ions provides confirmation of the nucleobase and the sequence. For potential de novo sequencing algorithms, it may be better to provide a product ion mass spectrum which only contains a single 5’- series and a single 3’-series of product ions (e.g., the c- and y-ions), but for database searching, more product ions (e.g., [a - Base], c-, w-, y-type ions) may yield more confident results. IRMPD of siRNAs can yield product ion mass spectra containing both types of information to improve confidence in the assignment of the siRNA sequence.
IRMPD vs. CID of Duplex siRNA
For effective down-regulation of target genes, duplex siRNA generally needs to be introduced to the organisms. The annealed duplex GAPDH and duplex Lamin A/C siRNAs were characterized by both CID and IRMPD. Strand separation was observed upon CID of duplex GAPDH in the 9− charge state as shown in Supplemental Figure 4a. Similar results were observed for the other charge states of duplex GAPDH and for duplex Lamin A/C (data not shown). Asymmetric charge partitioning of the duplex siRNAs is evident in Figure 7a in which the two single-strands are observed in the 6− charge state preferentially as opposed to the more symmetric 5− and 4− charge states. (It is presumed that the 3− charge state of the single-strands is also produced, however, these ions are not observed as they have m/z values beyond that of the mass analyzer.) The CID results are in line with previous reports on the gas-phase dissociation trends of DNA duplexes [53–55]. Diagnostic sequence ions were observed by CID when the collisional energy was increased as previously reported [31], but the sequence coverage was limited to less than 5 nucleotides (data not shown). IRMPD using 2.0 ms of irradiation yielded a product ion mass spectrum (Supplemental Figure 4b) almost identical to the CID mass spectrum in which the only dissociation channel observed was strand separation. With longer irradiation times, however, these single strands dissociated into diagnostic sequence ions, predominantly c- and y-type product ions, as shown in Supplemental Figure 4c. Due to homologous sequences between the two single strands, the origin of many of the product ions could not be unambiguously determined, and the overall sequence coverage of the individual single strands was low. All product ions observed were in the 1− or 2− charge state and none contained greater than eight nucleotides. While IRMPD of siRNA duplexes can yield informative product ions, the sequence information is limited to generally between 5 and 8 nucleotides from either terminal.
CONCLUSIONS
Infrared multiphoton dissociation of model single-strand siRNAs yields complete sequence coverage and has been shown to be a complementary dissociation method to traditional ion-trap based collision-induced dissociation. IRMPD affords rapid and efficient dissociation of siRNAs in less than 10 ms. The siRNA ions preferentially dissociated through cleavage of the 5’ P–O bonds by both IRMPD and CID to produce c/y-type sequence ions. The other dominant dissociation pathway was loss of neutral nucleobases from the siRNA anions or loss of protonated nucleobases from precursor cations. While CID produced high abundances of base loss ions, the irradiation time could be tuned to reduce the abundance of these uninformative product ions through secondary dissociation to yield higher abundances of c- and y-type fragment ions. The large degree of secondary dissociation also produced higher abundances of [a - Base] and w-type ions, as well as internal ions due to cleavage of two 5’ P–O bonds. At long irradiation times, larger and more highly charged product ions were also dissociated into shorter products in lower charge states. In fact, at irradiation times of greater than 10 ms, greater than 90% of the identified product ions were singly or doubly charged. While this effect reduced the total number of unique fragment ions in the IRMPD mass spectra, full sequence was still obtained and the product ion mass spectra was simpler to interpret. IRMPD of the siRNA cations produced high abundances of protonated bases with the exceptions of uracil and thymine. The protonated bases were only detected in the IRMPD mass spectra due to the alleviation of the low-mass cut-off, which would potentially allow one to confirm the presence and identity of chemically modified bases by performing IRMPD of RNA cations. Both CID and IRMPD of the siRNA cations in charge levels above 20% yielded full sequence coverage and only c- and y-type fragment ions were observed; [a - Base] and w-ions were detected at insignificant levels. Near complete sequence coverage was obtained for most of the siRNA anions and cations by both IRMPD and CID. The primary differences between the IRMPD and CID mass spectra were in the total number of product ions observed. With longer irradiation times, larger product ions were eliminated from the IRMPD mass spectra (due to conversion to smaller, diagnostic ions), however full sequence coverage was generally maintained in all cases. Short irradiation times typically yielded near complete series of c- and y-type ions, similar to results obtained by CID. These results suggest IRMPD analysis of siRNA in the positive ion mode may be a more fruitful means of sequencing oligoribonucleotides as the cations primarily dissociated through a single pathway to yield c- and y-type ions. Compared to IRMPD of siRNA anions in which product ions of all four types were observed, IRMPD of siRNA cations yields simplified mass spectra. RNA cations could potentially be de novo sequenced by IRMPD by identifying the c- and y-type ions corresponding to mass ladders of the 5’- and 3’-ends of the oligonucleotide, however, the irradiation time must be tuned to reduce the abundance of internal ions. While duplex siRNA predominantly underwent strand separation by CID, either single strands or sequence ions were produced by IRMPD by varying the irradiation time.
Supplementary Material
ACKNOWLEDGMENT
Teng-Yi Huang and Professor Scott McLuckey of Purdue University are gratefully acknowledged for their assistance with the siRNA desalting procedure. Funding from the Robert A. Welch Foundation (F-1155) and the National Institutes of Health (RO1 GM65956) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SYNOPSIS: Single-strand siRNA anions yield c- and y-type product ions as well as undergoing neutral base loss upon infrared multiphoton dissociation in a linear ion trap mass spectrometer.
SUPPLEMENTAL MATERIAL
Supplemental Figure 1 illustrates the IRMPD product ion mass spectra of [Lamin A/C.AS + 5H]5+ and [GAPDH.AS + 4H]4+. Supplemental Figures 2 and 3 show the charge state distribution of product ions and relative abundances of product ions of varying length as a function of irradiation time for GAPDH.S anions and cations, respectively. Supplemental Figure 4 shows the CID and IRMPD mass spectra of duplex GAPDH in the 9− charge state.
REFERENCES
- 1.Costa FF. Non-coding RNAs: New players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 2.Hüttenhofer A, Brosius J, Bachellerie J-P. RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Karagiannis TC, El-Osta A. RNA interference and potential therapeutic applications of short interfering RNAs. Cancer Gene Ther. 2005;12:787–795. doi: 10.1038/sj.cgt.7700857. [DOI] [PubMed] [Google Scholar]
- 5.Hudder A, Novak RF. miRNAs: Effectors of environmental influences on gene expression and disease. Toxicol. Sci. 2008;103:228–240. doi: 10.1093/toxsci/kfn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uprichard SL. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimm D. Small silencing RNAs: State-of-the-art. Adv. Drug Delivery Rev. 2009;61:672–703. doi: 10.1016/j.addr.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Roehl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher H-P, MacLachlan I. RNAi-mediated gene silencing in nonhuman primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 9.Miller VM, Gouvion CM, Davidson BL, Paulson HL. Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32:661–668. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YP, Haasnoot J, Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YP, Haasnoot J, ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limbach PA. Indirect mass spectrometric methods for characterizing and sequencing oligonucleotides. Mass Spectrom. Rev. 1997;15:297–336. doi: 10.1002/(SICI)1098-2787(1996)15:5<297::AID-MAS2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Hüttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedlaender MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 15.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller R-U, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter H-I, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozenski J, Vastmans K, Van Aerschot A, Herdewijn P. Evaluation of Capillary HPLC/Mass Spectrometry as an Alternative Analysis Method for Gel Electrophoresis of Oligonucleotides. Nucleosides, Nucleotides Nucleic Acids. 2003;22:1513–1516. doi: 10.1081/NCN-120023023. [DOI] [PubMed] [Google Scholar]
- 17.Suh M-J, Pourshahian S, Limbach PA. Developing Limited Proteolysis and Mass Spectrometry for the Characterization of Ribosome Topography. J. Am. Soc. Mass Spectrom. 2007;18:1304–1317. doi: 10.1016/j.jasms.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007;13:295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng Z, Limbach PA. RNase mapping of intact nucleic acids by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI-FTICRMS) and 18O labeling. Int. J. Mass Spectrom. 2004;234:37–44. [Google Scholar]
- 20.Hossain M, Limbach PA. Multiple endonucleases improve MALDI-MS signature digestion product detection of bacterial transfer RNAs. Anal. Bioanal. Chem. 2009;394:1125–1135. doi: 10.1007/s00216-008-2562-2. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 22.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 23.Little DP, Aaserud DJ, Valaskovic GA, McLafferty FW. Sequence Information from 42-108-mer DNAs (Complete for a 50-mer) by Tandem Mass Spectrometry. J. Am. Chem. Soc. 1996;118:9352–9359. [Google Scholar]
- 24.Keller KM, Brodbelt JS. Collisionally activated dissociation and infrared multiphoton dissociation of oligonucleotides in a quadrupole ion trap. Anal. Biochem. 2004;326:200–210. doi: 10.1016/j.ab.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Gabelica V, Tabarin T, Antoine R, Rosu F, Compagnon I, Broyer M, De Pauw E, Dugourd P. Electron Photodetachment Dissociation of DNA Polyanions in a Quadrupole Ion Trap Mass Spectrometer. Anal. Chem. 2006;78:6564–6572. doi: 10.1021/ac060753p. [DOI] [PubMed] [Google Scholar]
- 26.Smith SI, Brodbelt JS. Electron transfer dissociation of oligonucleotide cations. Int. J. Mass Spectrom. 2009;283:85–93. doi: 10.1016/j.ijms.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang T, Kharlamova A, Liu J, McLuckey SA. Ion Trap Collision-Induced Dissociation of Multiply Deprotonated RNA: c/y-Ions versus (a-B)/w-Ions. J. Am. Soc. Mass Spectrom. 2008;19:1832–1840. doi: 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Schürch S, Bernal-Mendez E, Leumann CJ. Electrospray tandem mass spectrometry of mixed-sequence RNA/DNA oligonucleotides. J. Am. Soc. Mass Spectrom. 2002;13:936–945. doi: 10.1016/S1044-0305(02)00413-0. [DOI] [PubMed] [Google Scholar]
- 29.Kirpekar F, Krogh TN. RNA fragmentation studied in a matrix-assisted laser desorption/ionisation tandem quadrupole/orthogonal time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2001;15:8–14. doi: 10.1002/1097-0231(20010115)15:1<8::AID-RCM185>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 30.Tromp JM, Schürch S. Gas-Phase Dissociation of Oligoribonucleotides and their Analogs Studied by Electrospray Ionization Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:1262–1268. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 31.Huang T, Liu J, Liang X, Hodges BDM, McLuckey SA. Collision-Induced Dissociation of Intact Duplex and Single-Stranded siRNA Anions. Anal. Chem. 2008;80:8501–8508. doi: 10.1021/ac801331h. [DOI] [PubMed] [Google Scholar]
- 32.Meng Z, Limbach PA. Shotgun sequencing small oligonucleotides by nozzle-skimmer dissociation and electrospray ionization mass spectrometry. Eur. J. Mass Spectrom. 2005;11:221–229. doi: 10.1255/ejms.736. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Håkansson K. Fragmentation of Oligoribonucleotides from Gas-Phase Ion-Electron Reactions. J. Am. Soc. Mass Spectrom. 2006;17:1369–1375. doi: 10.1016/j.jasms.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Mueller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, Dean NM. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emmerechts G, Barbe S, Herdewijn P, Anne J, Rozenski J. Post-transcriptional modification mapping in the Clostridium acetobutylicum 16S rRNA by mass spectrometry and reverse transcriptase assays. Nucleic Acids Res. 2007;35:3494–3503. doi: 10.1093/nar/gkm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmerechts G, Maes L, Herdewijn P, Anne J, Rozenski J. Characterization of the posttranscriptional modifications in Legionella pneumophila small-subunit ribosomal RNA. Chemistry & Biodiversity. 2008;5:2640–2653. doi: 10.1002/cbdv.200890218. [DOI] [PubMed] [Google Scholar]
- 38.Yu E, Fabris D. Direct Probing of RNA Structures and RNA-Protein Interactions in the HIV-1 Packaging Signal by Chemical Modification and Electrospray Ionization Fourier Transform Mass Spectrometry. J. Mol. Biol. 2003;330:211–223. doi: 10.1016/s0022-2836(03)00589-8. [DOI] [PubMed] [Google Scholar]
- 39.Kellersberger KA, Yu E, Kruppa GH, Young MM, Fabris D. Top-Down Characterization of Nucleic Acids Modified by Structural Probes Using High-Resolution Tandem Mass Spectrometry and Automated Data Interpretation. Anal. Chem. 2004;76:2438–2445. doi: 10.1021/ac0355045. [DOI] [PubMed] [Google Scholar]
- 40.Yu E, Fabris D. Toward multiplexing the application of solvent accessibility probes for the investigation of RNA three-dimensional structures by electrospray ionization-Fourier transform mass spectrometry. Anal. Biochem. 2004;334:356–366. doi: 10.1016/j.ab.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 41.Turner KB, Yi-Brunozzi HY, Brinson RG, Marino JP, Fabris D, Le Grice SFJ. SHAMS: combining chemical modification of RNA with mass spectrometry to examine polypurine tract-containing RNA/DNA hybrids. RNA. 2009;15:1605–1613. doi: 10.1261/rna.1615409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahr U, Aygun H, Karas M. Sequencing of Single and Double Stranded RNA Oligonucleotides by Acid Hydrolysis and MALDI Mass Spectrometry. Anal. Chem. 2009;81:3173–3179. doi: 10.1021/ac900100x. [DOI] [PubMed] [Google Scholar]
- 43.Sannes-Lowery KA, Hofstadler SA. Sequence confirmation of modified oligonucleotides using IRMPD in the external ion reservoir of an electrospray ionization fourier transform ion cyclotron mass spectrometer. J. Am. Soc. Mass Spectrom. 2003;14:825–833. doi: 10.1016/S1044-0305(03)00335-0. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, Håkansson K. Characterization of oligodeoxynucleotide fragmentation pathways in infrared multiphoton dissociation and electron detachment dissociation by Fourier transform ion cyclotron double resonance. Eur. J. Mass Spectrom. 2009;15:293–304. doi: 10.1255/ejms.966. [DOI] [PubMed] [Google Scholar]
- 45.Hofstadler SA, Griffey RH, Pasa-Tolic L, Smith RD. The use of a stable internal mass standard for accurate mass measurements of oligonucleotide fragment ions using electrospray ionization fourier transform ion cyclotron resonance mass spectrometry with infrared multiphoton dissociation. Rapid Commun. Mass Spectrom. 1998;12:1400–1404. [Google Scholar]
- 46.Silverstein RM, Webster FX, Kiemle DJ. Spectrometric Identification of Organic Compounds. 7th ed. Hoboken, NJ: John Wiley & Sons; 2005. [Google Scholar]
- 47.Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW. Infrared Multiphoton Dissociation of Large Multiply Charged Ions for Biomolecule Sequencing. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 48.Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Chromogenic Cross-Linker for the Characterization of Protein Structure by Infrared Multiphoton Dissociation Mass Spectrometry. Anal. Chem. 2008;80:4807–4819. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- 49.Lee JK. Insights into nucleic acid reactivity through gas-phase experimental and computational studies. Int. J. Mass Spectrom. 2005;240:261–272. [Google Scholar]
- 50.Liu M, Li T, Amegayibor FS, Cardoso DS, Fu Y, Lee JK. Gas-Phase Thermochemical Properties of Pyrimidine Nucleobases. J. Org. Chem. 2008;73:9283–9291. doi: 10.1021/jo801822s. [DOI] [PubMed] [Google Scholar]
- 51.Zhachkina A, Liu M, Sun X, Amegayibor FS, Lee JK. Gas-Phase Thermochemical Properties of the Damaged Base O6-Methylguanine versus Adenine and Guanine. J. Org. Chem. 2009;74:7429–7440. doi: 10.1021/jo901479m. [DOI] [PubMed] [Google Scholar]
- 52.Pan S, Verhoeven K, Lee JK. Investigation of the Initial Fragmentation of Oligodeoxynucleotides in a Quadrupole Ion Trap: Charge Level-Related Base Loss. J. Am. Soc. Mass Spectrom. 2005;16:1853–1865. doi: 10.1016/j.jasms.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Gabelica V, De Pauw E. Comparison between solution-phase stability and gas-phase kinetic stability of oligodeoxynucleotide duplexes. J. Mass Spectrom. 2001;36:397–402. doi: 10.1002/jms.141. [DOI] [PubMed] [Google Scholar]
- 54.Gabelica V, De Pauw E. Comparison of the collision-induced dissociation of duplex DNA at different collision regimes: Evidence for a multistep dissociation mechanism. J. Am. Soc. Mass Spectrom. 2002;13:91–98. doi: 10.1016/s1044-0305(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 55.Wan KX, Gross ML, Shibue T. Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J. Am. Soc. Mass Spectrom. 2000;11:450–457. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.