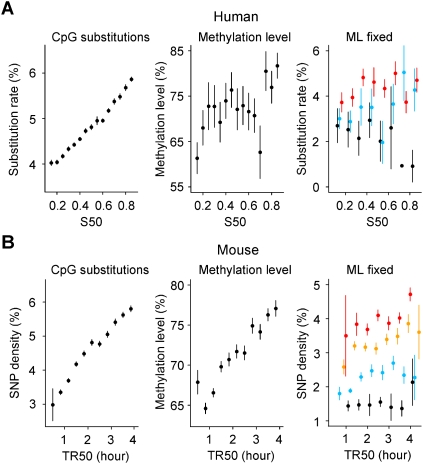
Figure 5.
Increase of human and mouse CpG substitution rates in later-replicating regions explained by the increase in methylation level. Replication timing and CpG substitution rates are determined in noncoding regions excluding CpG islands (Methods). (A, left) Human CpG substitution rate as function of S50; (center) methylation level determined in human sperm cells (Eckhardt et al. 2006) plotted as a function of the replication timing S50; (right) human CpG substitution rate when controlling for methylation level (ML). (Black) ML ≤ 20%; (blue) 20% < ML ≤ 60%; (red) ML > 60%. (B) Analyses performed with mouse replication timing data TR50 (Farkash-Amar et al. 2008). (Left) Mouse CpG diversity was computed with SNP data (The International HapMap Consortium 2007); (center) methylation level determined in mouse embryonic stem cells (Meissner et al. 2008) plotted as a function of replication timing; (right) Mouse CpG SNP density when controlling for ML. (Black) ML ≤ 45%; (blue) 45% < ML ≤ 60%; (orange) 60% < ML ≤ 70%; (red) ML > 70%. DNA methylation levels, substitution rates, divergence, and replication timing were computed as indicated in Methods. The window size is as indicated in Methods (DNA Methylation section).