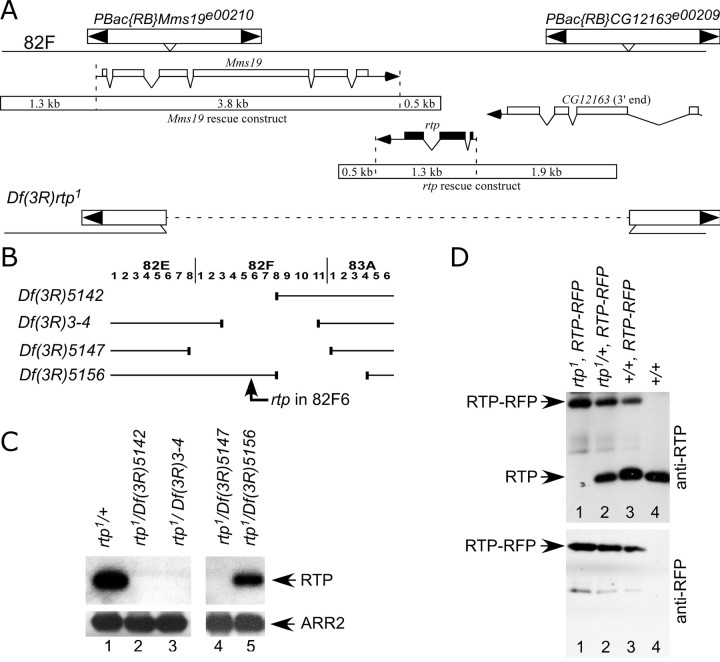
Figure 4.
Creation and analysis of the rtp1 mutant. A, The genomic region that contains CG10233 (rtp) is flanked by the PBac insertions PBac{RB}Mms19e00210 and PBac{RB}CG12163e00209. FLP-directed recombination between these insertions produced a deletion chromosome, Df(3R)rtp1, that deleted CG10233 and disrupted the Mms19 and CG12163 genes. The 5.6 kb genomic DNA fragment containing the Mms19+ gene (Mms19 rescue construct) that rescued the zygotic lethality of Df(3R)rtp1 and the 3.7 kb genomic DNA used in the rtp rescue construct are also shown. B, Diagram showing the four deficiency chromosomes used in the analysis of the rtp gene. The location of the rtp gene in 82F6 is indicated. The rtp gene was missing in all deletions except Df(3R)5156. C, Top panels, Protein blots show that rtp/+ and rtp1/Df 5156 retain RTP protein, but RTP protein was absent in all rtp1/Df genotypes expected to lack the rtp gene. Bottom panels, Reprobing the protein blot with ARR2 antibodies demonstrated that similar levels of retinal proteins were loaded in all lanes. D, Protein blot analysis of the RTP-RFP fusion protein. The RTP-RFP 50 kDa protein was identified by both anti-RTP and anti-RFP antibodies (lanes 1–3). The levels of RTP-RFP protein was greatest when no native RTP is expressed (lane 1) but diminished in a dose-responsive manner when one copy (lane 2) or two copies (lane 3) of the native RTP were also expressed.