Figure 2.
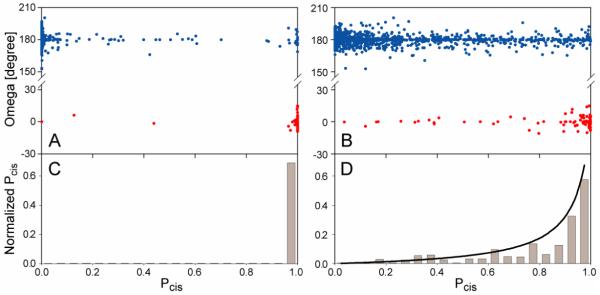
Prediction of Xaa-Pro peptide bond conformation from NMR chemical shifts by the program Promega. (A,B) Plot of the crystallographically observed ω angles, versus their probability score, Pcis, calculated by eq 5, (A) for residues that included 13Cγ chemical shift assignments, and (B) for all database residues with at least three out of four (13C’, 13Cα, 13Cβ, 1Hα) Pro assignments, and ignoring 13Cγ chemical shifts. Note that Pcis (eq 5) corresponds to a relative probability, i.e., Pcis = 0.5 refers to the case where the cis probability equals the average cis probability in the database (ca 5%). (C,D) Histograms of the normalized, true probability score , calculated by using eq 6 for Pro residues in the database in (C) the presence and (D) the absence of 13Cγ chemical shifts. The solid line in (D) corresponds to eq 6. Note that in the presence of 13Cγ chemical shifts the Pcis result is essentially binary, and the below unity value observed in the histogram for the highest Pcis bin largely reflects cases where solution and crystal structures differ.