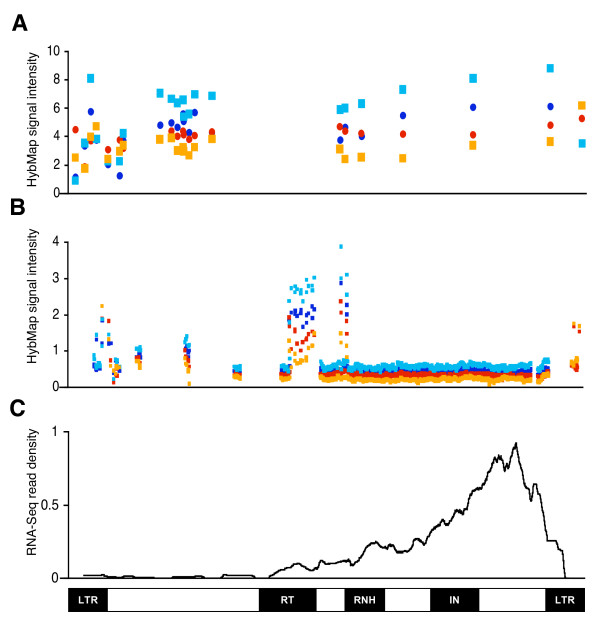
Figure 3.
Transcriptional activity along full-length LTR retrotransposons. A) Signal intensities of HybMap probes (y-axis) mapping uniquely to a single full-length LTR locus are plotted along the retrotransposon sequence (x-axis). Total RNA samples are shown as blue circles (forward strand probes) and red circles (reverse strand probes). Poly(A)-enriched samples are shown as light blue squares (forward) and orange squares (reverse). B) As above, but with probes mapping to multiple full-length LTR retrotransposons. C) The density of RNA-Seq reads mapping to the LTRs are shown as the average using a 301-bp long sliding window. Density shown as reads mapped per sequence in the alignment. The genetic structure of an LTR retrotransposon is depicted at the bottom, with flanking LTR sequences, as well as reverse transcriptase (RT), RNaseH (RNH) and integrase (IN) domains indicated.