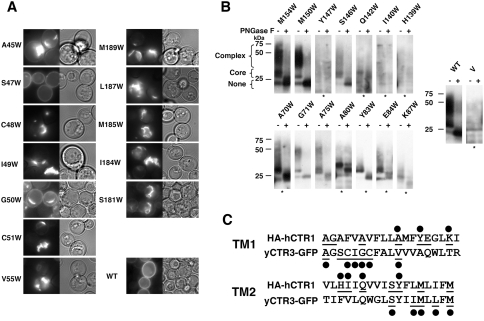
Fig. 2.
Trp mutations cause processing defects in both TM1 and TM2. a Yeast transformed with WT yCTR3-GFP and Trp mutants that displayed a null complementation phenotype were subjected to fluorescence microscopy. For each indicated mutant an image was captured in fluorescent (left) and DIC (right) mode. b Crude membranes of yeast transformed with nonfunctional HA-hCTR1 Trp mutants were resuspended in buffer, normalized for total protein and incubated in the absence or presence (– and +, respectively) of PGNase F to remove N-linked glycosylation. Samples were then subjected to Western blotting for the α-HA epitope. Since the levels of expression varied between mutants, some of the blots needed to be overexposed relative to the WT (*). In addition to the relative level of expression, blots revealed whether the mutants retained any ability to acquire complex N-glycosylation patterns that would indicate exit of the protein from the ER (see brackets). A negative control sample from cells just carrying the vector is labeled V. c Summary of positions whose posttranslational processing and/or trafficking was impaired. As in Fig. 1, all residues assayed in this experiment are underlined and those that were sensitive to Trp are marked with a black circle