Figure 2.
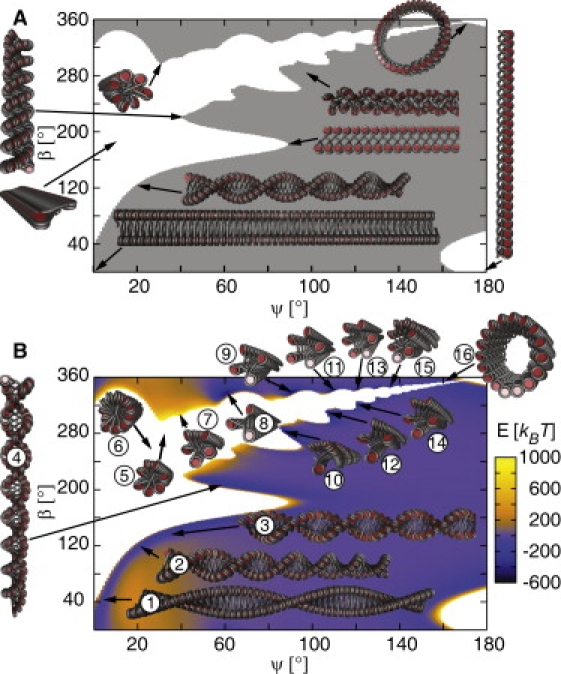
Static energy phase diagrams of the CLS geometry with NRL = 212 bp for the opening angle ψ (γ = 0) and the nucleosome twist angle β with a step width of 1°. Fiber conformations with sterical overlaps are represented in white. (A) A classical phase diagram representation in which the region of fiber conformations without sterical overlaps is depicted in gray. (B) A phase diagram showing the energy E associated with a given fiber conformation. The diagram exhibits regions of very high energy caused by electrostatic DNA repulsion. These include fibers with crossed-linker DNA, e.g., [5, 2], [11, 5] and [7, 3] (fibers 5–7). In contrast, [2, 1] zig-zag fiber conformations (fibers 1–4) and [3, 1] and [4, 1] fibers (fibers 8–10) have the broadest energy valleys, whereas [n, 1] structures with n > 4 exhibit lower energy values but smaller valleys (fibers 11–16). Fiber 6 represents our previous model of chromatin from chicken erythrocytes (25).
