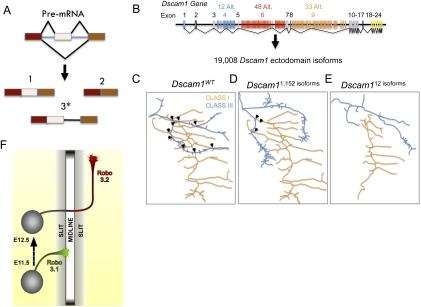
Figure 2.
Roles of AS in neuronal connectivity. (A) Forms of AS. Most eukaryotic genes consist of protein-coding exon (rectangles) and noncoding intron (lines) sequences. Following transcription, the splicesome and splicing factors mediate intron removal from pre-mRNA to create a mature mRNA transcript. AS is a mechanism by which exons are included or excluded from the mRNA (1,2), thereby creating isoforms (also called variants) of distinct protein structures. In some instances, AS may result in differential intron retention (3*), such as in the case of Robo3 (Chen et al. 2008), which generates a premature stop codon that vastly changes the C-terminal of the encoded gene. (B) Gene structure of Dscam1 ectodomains illustrating the potential AS isoforms. (C–E) Representative illustrations of dendritic arborization neurons in different Dscam1 mutant backgrounds (arrows represent dendritic branch overlap). (C) Dendrites of different dendritic arborization classes overlap extensively. (D) However, in Dscam1 mutants expressing 1152 isoforms, the amount of overlap between class I and III dendrites decreased significantly. (E) When Dscam1 diversity decreased to 12 or 24, dendrites of distinct classes completely avoid each other, indicating that appropriate patterning of dendritic arborization neurons depends on thousands of Dscam1 isoforms (Hattori et al. 2009). (F) Schematic of a commissural neuron during embryonic development as seen in an open book spinal cord dissection. Axonal shafts represent the differential expression patterns of Robo3 isoforms. In this context, Robo3.1 enables axons to cross the midline, whereas Robo3.2 prevents axons from recrossing (Chen et al. 2008).