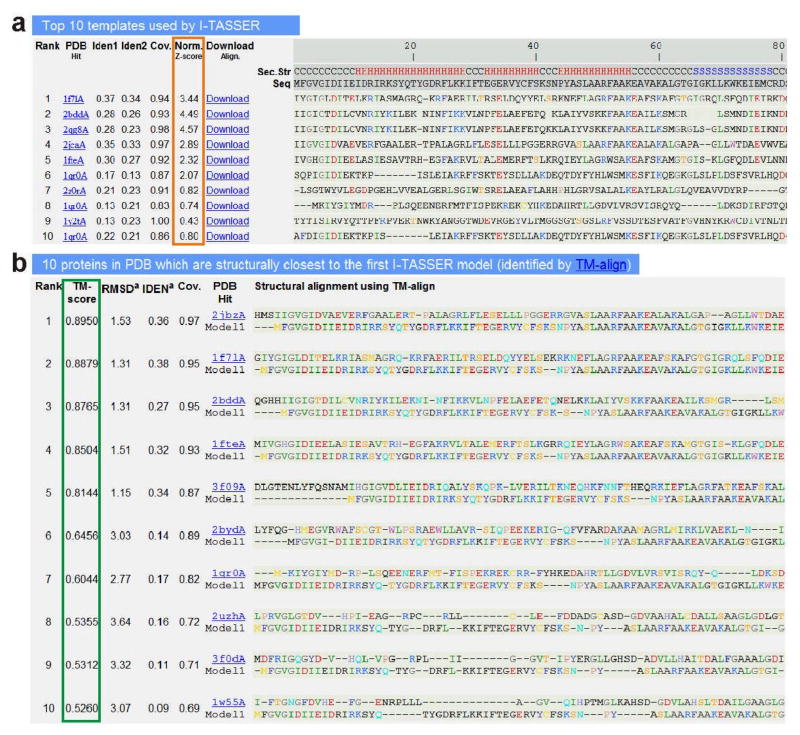
Figure 4.
An illustrative example of the I-TASSER result page showing (a) top ten threading templates and the alignments for the query protein identified by LOMETS; and (b) structural analogs and their alignment with the I-TASSER model, as identified by TM-align from the PDB library. The quality of the threading alignment in (a) is evaluated based on their normalized Z-score (highlighted in orange), where a normalized Z-score >1 reflects a good alignment. The ranking of the analogs shown in (b) is based on the TM-score (highlighted in green) of the structural alignment.