Abstract
Sponge bodies, cytoplasmic structures containing post-transcriptional regulatory factors, are distributed throughout the nurse cells and oocytes of the Drosophila ovary and share components with P bodies of yeast and mammalian cells. We show that sponge body composition differs between nurse cells and the oocyte, and that the sponge bodies change composition rapidly after entry into the oocyte. We identify conditions that affect sponge body organization. At one extreme, components are distributed relatively uniformly or in small dispersed bodies. At the other extreme, components are present in large reticulated bodies. Both types of sponge bodies allow normal development, but show substantial differences in distribution of Staufen protein and oskar mRNA, whose localization within the oocyte is essential for axial patterning. Based on these and other results we propose a model for the relationship between P bodies and the various cytoplasmic bodies containing P body proteins in the Drosophila ovary.
Keywords: sponge bodies, P bodies, oogenesis
Introduction
Mechanisms that regulate the translation or localization of mRNAs have long been recognized for their crucial roles in the early stages of development (Bashirullah et al., 1998; Johnstone and Lasko, 2001; Colegrove-Otero et al., 2005; King et al., 2005). More recently, there has been a growing awareness that post-transcriptional regulation is widespread, and contributes to the development or function of most if not all cells (Moore, 2005). Some of these control mechanisms act globally. For example, the overall translational capacity of a cell can be adjusted by modulating the activity of a rate-limiting factor (Sonenberg, 1996). Other mechanisms act on individual mRNAs, and thus require factors that recognize the mRNAs and provide specificity. Recognition can be mediated by proteins, or by small noncoding RNAs that bind specifically to mRNAs and target them for degradation or translational repression (Bartel, 2004; Colegrove-Otero et al., 2005). Independent of what type of molecule serves to recognize a regulated mRNA, other factors must come into play to degrade, sequester, repress, activate, move or modify the transcripts. Classically, the identity and action of these additional factors has been explored largely through biochemical or genetic approaches, with an emphasis on defining biochemical reactions and establishing in vitro assays in which regulation can be recapitulated. More recently, much attention has turned to the sites within cells where regulation occurs.
Studies of mRNA turnover in yeast led to the discovery of P bodies, large RNPs that are devoid of ribosomes (Teixeira et al., 2005). Initially, P bodies were found to contain proteins with roles in mRNA degradation, and only those mRNAs destined for turnover (Bashkirov et al., 1997; Ingelfinger et al., 2002; van Dijk et al., 2002; Eystathioy et al., 2003; Sheth and Parker, 2003). Progress in understanding these large RNPs has revealed a broader role. Not all transcripts in P bodies are on the path to decay, and some fraction of mRNAs can later be translated (Brengues et al., 2005; Bhattacharyya et al., 2006). Thus the P bodies are also sites for storage and repression of mRNAs. Components of P bodies are highly conserved and have led to the identification of similar or equivalent particles in a wide variety of species, including Drosophila (Ingelfinger et al., 2002; Lykke-Andersen, 2002; van Dijk et al., 2002; Sheth and Parker, 2003; Cougot et al., 2004).
The sponge bodies of Drosophila ovaries provide another example of a subcellular structure correlated with post-transcriptional regulation. The Drosophila egg chamber includes 16 sister germline cells - 15 nurse cells and a single oocyte - which, owing to incomplete cytokinesis, remain interconnected by ring canals (Spradling, 1993). The nurse cells are transcriptionally active and produce most or all of the transcripts deposited in the oocyte and provided maternally to the embryo. Sponge bodies appear in the nurse cells and oocyte, and consist of endoplasmic reticulum (ER)-like cisternae or vesicles embedded in an electron dense matrix that excludes most ribosomes. They were first characterized because of their enrichment for Exuperantia (Exu) (Wilsch-Brauninger et al., 1997), a protein acting in localization of bicoid (bcd) and other mRNAs to specific sites within the oocyte (Berleth et al., 1988; St Johnston et al., 1989; Wilhelm et al., 2000). Sponge bodies can also be monitored by fluorescence microscopy, using Exu∷GFP as a marker (Wang and Hazelrigg, 1994). At this level of resolution they appear as cytoplasmic particles, some of which approach or abut the nucleus. Several additional sponge body proteins have been identified on the basis of their colocalization with Exu∷GFP using fluorescence microscopy. Ultrastructural studies of one of these, Me31B, confirms that it is localized to sponge bodies (Nakamura et al., 2001), providing additional evidence that colocalization of proteins in the cytoplasmic particles as measured by fluorescence microscopy is a reliable indicator of sponge body association. Notably, most if not all of the sponge body proteins have been implicated in post-transcriptional control of gene expression (see Table 1, below), supporting the view that sponge bodies are sites through which regulated mRNAs traffic or are stored. Some of these proteins are the homologs of P body components, which has led to suggestions that sponge bodies and P bodies are related, and even to interchangeable use of the names (Decker and Parker, 2006; Lin et al., 2006).
Table 1.
Sponge body associated proteins.
| Protein | Localized mRNAs that are regulated | sponge body localization |
|---|---|---|
| Exu | bcd 1, osk 2 | 25 |
| Btz | osk 3 | 5 |
| Cup | osk 4,5, nos 6 | 4,5 |
| eIF4E | osk 4,5 | 4 |
| Me31B | osk 7 | 7 |
| Yps | osk?2,8,9 | 2 |
| Gus | 10 | 10 |
| Dcp1 | osk11 | 11 |
| Dcp2 | 11 | |
| Sqd | osk 12, grk 13,14 | 15 |
| BicC | osk16 | 17 |
| Hrb27C | osk 18,19, grk 20 | this study and unpublished in 15 |
| Bru | osk 21, grk 22,23 | this study |
| Orb | osk 24,25, grk 26 | this study |
(Wilsch-Brauninger et al., 1997); 3 (van Eeden et al., 2001);
(Nelson et al., 2004);
(Mansfield et al., 2002);
(Styhler et al., 2002);
(Norvell et al., 2005);
(Kelley, 1993);
(Norvell et al., 1999);
(Saffman et al., 1998);
(Huynh et al., 2004);
(Yano et al., 2004);
(Goodrich et al., 2004);
(Webster et al., 1997);
(Filardo and Ephrussi, 2003);
(Yan and Macdonald, 2004);
(Chang et al., 1999);
(Castagnetti and Ephrussi, 2003);
Here we further characterize the composition of sponge bodies, identifying additional regulatory factors that co-localize with Exu∷GFP in nurse cells or the oocyte. We also show that sponge body organization is dynamic: composition changes rapidly after transit from nurse cells to the oocyte. In addition, sponge body organization is dependent on environmental conditions, and changes have dramatic effects on the distribution of oskar (osk) mRNA during the process of its localization to the posterior pole of the oocyte. The dynamic nature and plasticity of sponge bodies suggests a model for the hierarchical organization of ribonucleoprotein (RNP) particle assemblies in the ovary.
Results
Identification of new sponge body components
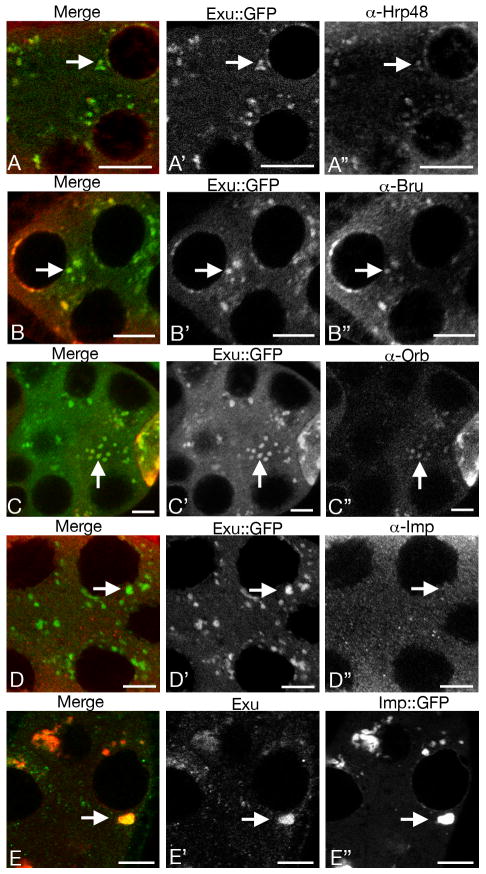
Many proteins with roles in post-transcriptional regulation are associated with sponge bodies. To identify additional sponge body components we tested a panel of post-transcriptional regulatory factors and identified four – BicC (Snee and Macdonald, submitted for publication), Hrb27C (also known as Hrp48), Bru, and Orb – that are extensively colocalized with Exu∷GFP (Table 1 and Fig. 1). Two other regulatory factors tested, Bicoid Stabilization Factor (Mancebo et al., 2001) and Imp (Nielsen et al., 2000; Geng and Macdonald, 2006; Munro et al., 2006), are not enriched in sponge bodies (Fig. 1D and data not shown). The absence of Imp in sponge bodies is at odds with a recent report in which a form of Imp tagged with GFP was observed in large particles in the ovary (Boylan et al., 2008). To explore this discrepancy, the distribution of an Imp∷GFP fusion protein was tested. Unlike Imp, Imp∷GFP is highly enriched in sponge bodies (Fig. 1E). Thus, tagging with GFP appears to drive Imp into sponge bodies.
Fig. 1. Bru, Orb and Hrp48 are associated with sponge bodies.
All panels are of stage 7 or 8 egg chambers. In each row the black and white panels (e.g. A′ and A″) show the distributions of proteins as indicated, and the left panel (e.g. A) shows a color merge in which the signals from the center and right panels appear in green and red, respectively. Representative foci of sponge bodies are indicated by arrows. Although Imp is not detectably concentrated in sponge bodies (D), an Imp∷GFP fusion protein expressed from a transgene is (E). Scale bars are 10 μm.
Another approach to identify candidate sponge body factors, which does not rely on the availability of antibodies, involves the use of GFP traps. GFP traps are fly strains in which a GFP-bearing exon has been transposed into an intron, leading to the expression of a GFP fusion protein. Published images of GFP distribution in the GFP traps can be examined to find those of interest (Morin et al., 2001; Kelso et al., 2004). Sponge bodies have a characteristic distribution, appearing as cytoplasmic granular material that is often associated with the outer surface of nurse cell nuclei. Three GFP trap lines (of those available in the original collection (Morin et al., 2001; Kelso et al., 2004)) had such an appearance and were selected for further analysis.
One GFP trap was reported as an insertion in the CG3634 gene, which encodes a protein of unknown function. However, we found that this fly strain contained two GFP trap insertions. The second is in the me31B gene, which encodes a sponge body component (Nakamura et al., 2001); this is the trap responsible for the sponge body GFP pattern (Materials and methods). The Me31B∷GFP trap, like Exu∷GFP, is useful as a sponge body marker for colocalization studies.
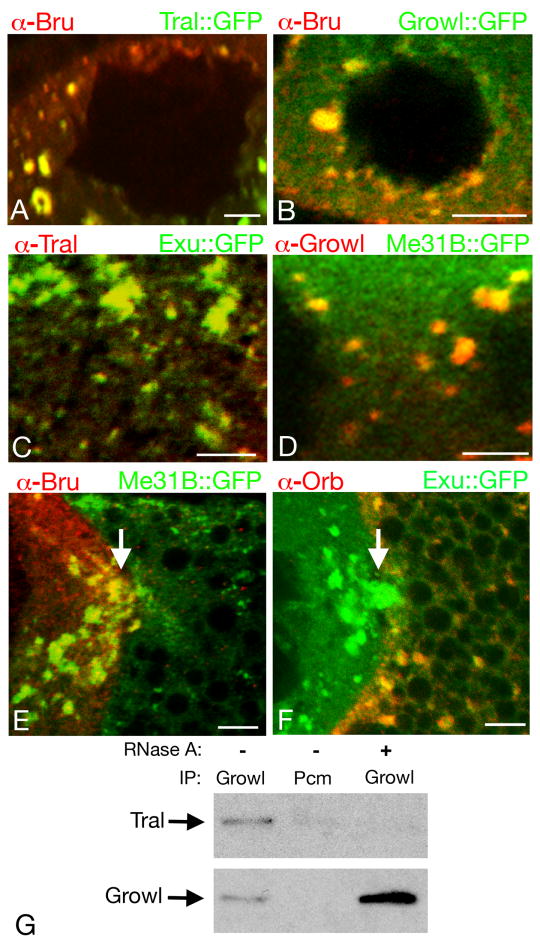
The other selected GFP traps are in the trailer hitch (tral) and growl (CG14648) genes. Double labeling experiments with immunodetection of Bru protein confirm that both GFP traps label sponge bodies (Fig. 2A,B). Although the GFP trap proteins are expected to reveal the distribution of the endogenous, untagged protein, confirmation by immunodetection of the native (untagged) proteins is necessary. The Tral protein has already been shown to colocalize with sponge body proteins (Wilhelm et al., 2005), and we have confirmed colocalization with Exu∷GFP (Fig. 2C). Native Growl protein is also expressed in the ovary (supplementary material Fig. S1) where it is cytoplasmic and colocalizes with a sponge body marker (Fig. 2D). Thus, using a simple screening criterion – particulate cytoplasmic distribution of GFP in images provided for the entire collection of GFP traps – it is possible to identify proteins that are strong candidates for sponge body components.
Fig. 2. Sponge body composition and remodeling.
Panels A-D show portions of nurse cells from stage 7 egg chambers. For panels A and B, Bru (red) was detected by immunostaining, and Tral∷GFP (A) and Growl∷GFP (B) (both green) were detected by GFP fluorescence. In each case Bru and the GFP fusion protein are both enriched, and colocalize, in sponge bodies. The circular regions devoid of staining are nurse cell nuclei. For panels C and D the endogenous Tral (C) and Growl (D) proteins were detected by immunostaining (red), with GFP fusion proteins to mark sponge bodies (Exu∷GFP in C, Me31B∷GFP in D). Both Tral and Growl are highly enriched in sponge bodies. Regions devoid of staining in D are portions of nurse cell nuclei. Panel C does not include a nurse cell nucleus, but Tral is not nuclear (Wilhelm et al., 2005).
Panels E and F show portions of stage 9 egg chambers including the boundary between the oocyte (at right) and a nurse cell (at left). In E, Bru (red) is present at high levels in the nurse cell sponge bodies (the arrow shows an example), but is almost completely undetectable in the oocyte sponge bodies, most of which are small and dispersed in this egg chamber. The sponge bodies are marked with Me31B∷GFP. In F, Orb (red) cannot be detected in the nurse cell sponge bodies, but is present at high levels in the oocyte sponge bodies. The sponge bodies are marked with Exu∷GFP. The positions of ring canals connecting the nurse cells and oocytes are indicated by arrows. Scale bars are 10 μm in all panels.
Panel G shows the RNA dependent interaction between Tral and Growl. Immunoprecipitations were performed out of ovarian extracts using Growl or a negative control antibody (Pcm) in the presence or absence of RNase A. Western blots of the immunoprecipitated proteins were performed using anti-Tral antibody (top panel) or Growl antibody (bottom panel).
In addition to being concentrated in sponge bodies, Tral and Growl proteins can be coimmunoprecipitated (Fig. 2G). The interaction is sensitive to RNase treatment, indicating that the proteins are bound to common RNAs and are not engaged in direct contacts, or that the proteins bind directly to one another in a manner dependent on RNA (Fig. 2G).
Rapid reorganization of sponge bodies upon entry into the oocyte
The composition of sponge bodies differs between nurse cells and the oocyte. Although some components, such as Me31B and Exu, are present at similar levels in both cell types, other sponge body proteins can be highly enriched in either nurse cells or the oocyte. For example, in vitellogenic egg chambers Bru is concentrated in the nurse cells, while Orb is concentrated in the oocyte. This partitioning is observed both for the fraction of each protein that colocalizes with sponge bodies, and for the fraction that is spread more uniformly throughout the cytoplasm (Fig. 2E,F).
Sponge bodies are not fixed in position, and move via transit through ring canals between different nurse cells as well as unidirectionally from nurse cells into the oocyte (Theurkauf and Hazelrigg, 1998). To begin to explore the process by which sponge body composition changes after movement into the oocyte, we wished to determine how far sponge bodies travel into the oocyte before they lose Bru and gain Orb. Me31B∷GFP and Exu∷GFP were used to track the sponge bodies in stage 8 and 9 egg chambers, and Bru and Orb were detected by immunofluorescence. Oocyte sponge bodies immediately adjacent to ring canals were scored as positive or negative for intense Bru or Orb and their distance from ring canals was determined. The furthest Bru-positive sponge body was 5.6 μm from the ring canals and the closest Bru-negative sponge body was 2.4 μm from the ring canals (22 sponge bodies analyzed). The furthest Orb-negative sponge body was 3.7 μm from ring canals and the closest Orb-positive sponge body was 2.4 μm from ring canals (9 sponge bodies analyzed). Thus, reorganization of the sponge bodies, at least in terms of elimination of Bru and recruitment of Orb, occurs within a narrow transition zone positioned approximately 2-6 μm away from the ring canals. Considering that most sponge bodies move at a rate of 6-8 μm/min through ring canals (Theurkauf and Hazelrigg, 1998), and that the rate of movement of sponge bodies in the transition zone appears to be the same ((Mische et al., 2007); data not shown), the sponge bodies presumably change composition within a minute after entering the oocyte.
Sponge body architecture changes in response to environmental conditions
During the course of our analysis, substantial variation was observed in the distribution of sponge body markers in ovaries from different females of the same genotype. Similarly, published images of sponge body protein distributions show considerable differences (Wilsch-Brauninger et al., 1997; Theurkauf and Hazelrigg, 1998; Wilhelm et al., 2000; Nakamura et al., 2001; Nakamura et al., 2004; Lin et al., 2006). Systematic tests of the possible causes of this variation, all performed with healthy flies that had not been subjected to overcrowding, revealed three sets of conditions that reliably yield only one of two extreme types in the spectrum of Me31B∷GFP distribution: in dispersed or reticulated bodies (each type is described in detail below). Dispersed bodies are consistently observed in essentially all egg chambers from 3-4 day old females that are provided with the standard cornmeal growth medium that has been supplemented with dried yeast on the surface of the medium. Addition of surface yeast is an approach widely used to enhance egg laying (King, 1970; Ashburner, 1989). Reticulated bodies, the second type, are consistently seen in essentially every egg chamber using the same rich medium but without the additional surface yeast. Reticulated bodies are also consistently found in virgin females, maintained on the rich medium with or without surface yeast. Other conditions yield a more heterogenous population of the different Me31B∷GFP distribution types (e.g. Fig. 2D,E).
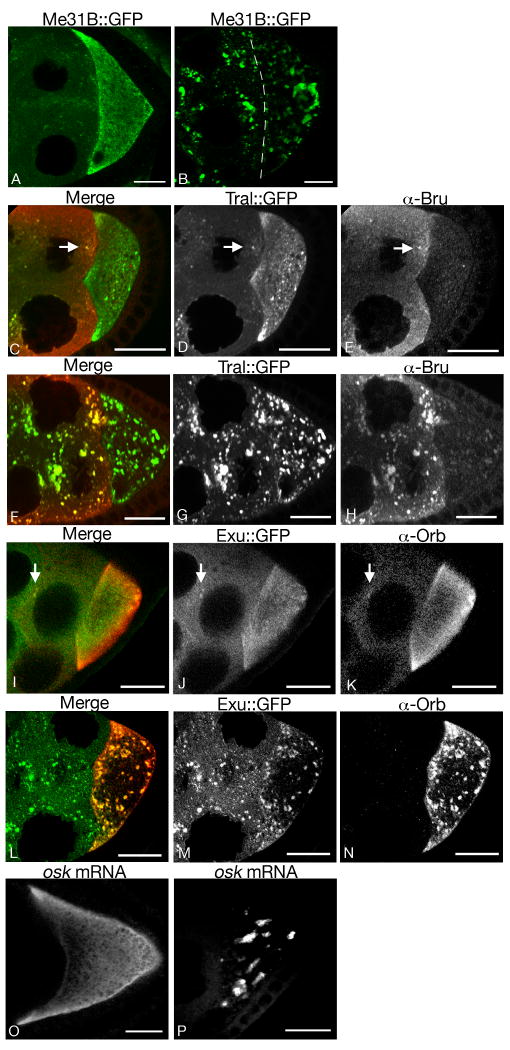
A typical example of dispersed bodies is shown in Fig. 3A, using live imaging and Me31B∷GFP as a marker. Me31B∷GFP appears in small puncta that are dispersed throughout the nurse cells and oocyte. Although Me31B∷GFP is clearly concentrated in the puncta, there is also a high level of the protein more evenly distributed throughout the cytoplasm. Similar patterns are observed for other sponge body markers, including Tral∷GFP, Exu∷GFP, Bru, and Orb (Fig. 3C-E, I-K). Some variation is observed for different proteins. For example, Me31B∷GFP, Tral∷GFP and Bru are present in many small puncta, while Exu∷GFP and Orb are more evenly distributed throughout the cytoplasm. Similar results were obtained using both live imaging (where possible) and fixed samples.
Fig. 3. Plasticity of sponge bodies.
Distribution of molecules in egg chambers from females maintained under conditions that promote formation of one of the two extreme forms of sponge bodies, either dispersed (A, C-E, I-K, O) or reticulated (B, F-H, L-N, P). The proteins being detected are indicated above. Each panel shows the oocyte (at right) and some of the nurse cells (at left). The nurse cell to oocyte boundary is not obvious in B, and is therefore marked with a dashed line.
Panels A and B show Me31B∷GFP fluorescence in live egg chambers. Note the significant reduction in diffuse cytoplasmic staining in B relative to A.
Panels C-H compare Tral∷GFP (D,G and green in C,F) and Bru (E,H and red in C,F). Both are concentrated in the small puncta of dispersed bodies (arrows in C-E) and in the many larger puncta of reticulated bodies. At this stage Bru is largely restricted to nurse cells, and the puncta in the oocyte are labeled exclusively with Tral∷GFP.
Panels I-N compare Exu∷GFP (J,M and green in I,L) and Orb protein (K,N and red in I,L). Both proteins are detected in the relatively rare small puncta of dispersed bodies (arrows), although the level of Orb is much lower in the nurse cells than in the oocyte. Both Exu∷GFP and Orb concentrate in reticulated bodies that form in the oocyte cytoplasm of stage 8 egg chambers.
Panels O and P show osk mRNA in stage 8 egg chambers. osk RNA before its localization to the posterior is either dispersed in the cytoplasm with concentration at the anterior margins when dispersed sponge bodies are present (O), or localizes to reticulated bodies when these structures form (P, see also Fig. 5D).
Scale bars are 20 mm in all panels.
Examples of reticulated bodies are shown in Fig. 3B, using live imaging and Me31B∷GFP as a marker. Reticulated bodies are much larger than dispersed bodies, and often appear to be extensively interconnected (see also Fig. 5M,N). The size and number of these bodies can vary in different samples, possibly due to environmental influences that we have been unable to control. Cytoplasmic levels of Me31B∷GFP are low (compared to conditions where dispersed bodies form), suggesting that the protein is recruited into reticulated bodies. Similar patterns are observed for other sponge body markers in reticulated bodies, including Tral∷GFP, Exu∷GFP, Bru, and Orb (Fig 3F-H, L-N). The reticulated bodies form in nurse cells and oocytes during stages 6-10B of oogenesis, but they are the largest and most reticular in the oocytes of stage 7–9 egg chambers.
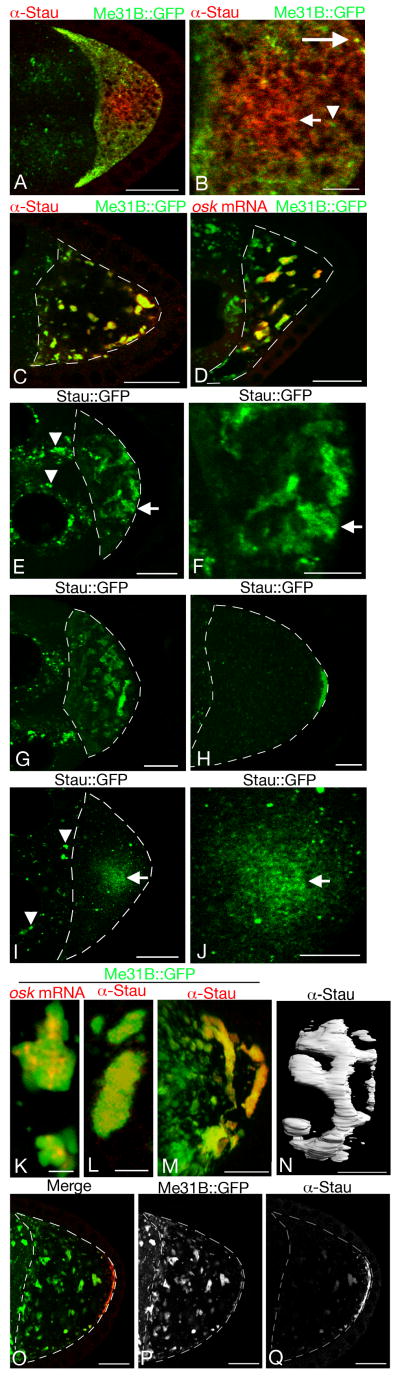
Fig. 5. Sponge body organization affects distribution of Stau protein and osk mRNA.
Panel A shows a portion of a stage 8 egg chamber with dispersed sponge bodies, with a higher magnification of the oocyte in panel B. Stau is red and Me31B∷GFP is green. Some of the sponge bodies in the oocyte contain Stau (the yellow puncta such as the one indicated by a long arrow), and others do not (green puncta such as that indicated by the arrowhead). Most of Stau, which is enriched in the oocyte relative to the nurse cells, appears in structures that do not contain high levels of Me31B∷GFP (short arrow). Panels C and D show stage 8 egg chambers with reticulated bodies labeled with Me31∷GFP (green) and stained for either Stau (C) or osk mRNA (D) in red (osk mRNA was detected by fluorescent in situ hybridization). Both Stau and osk mRNA are strongly concentrated in the reticulated bodies. The dashed lines in panels C-E, G-I, and O-Q trace the oocyte outline.
Panels E-J are of Stau∷GFP in live egg chambers (all other images in this figure are from fixed samples) in conditions with reticulated bodies (E-H) and dispersed bodies (I,J). Panels F and J are higher magnification views of E and I, respectively. As for endogenous Stau, Stau∷GFP is highly concentrated in reticulated bodies (arrows in E,F; early stage 8), is progressively enriched in bodies near the posterior of stage 8 oocytes (G), and is tightly localized to the posterior after stage 9 (H). In egg chambers with dispersed bodies (I,J), Stau∷GFP parallels the pattern of endogenous Stau shown in A and is cytoplasmic, but is also present in many small particles that are not seen for endogenous Stau in fixed tissue (arrows in I,J). J is a close up view of I and the arrows indicate the same cytoplasmic region in both images. Stau∷GFP localizes tightly to the posterior after stage 9 when dispersed bodies are present (data not shown) and its distribution is indistinguishable from that when reticular bodies are present (as in H). Examples of the artifactual class of Stau∷GFP particles, which do not correspond to endogenous Stau (see text), are indicated by arrowheads in E and I.
Panels K and L show the concentration of osk mRNA (red in K) and Stau (red in L) in subdomains of reticulated bodies labeled uniformly with Me31B∷GFP. Panel M and N are images from a 3D reconstruction of Stau (red in M, white in N) and Me31B∷GFP (green in M) in a stage 8 egg chamber. Reticulated bodies appear to form a highly interconnected network with Stau enriched at the posterior (M). N shows the highly interconnected nature of Stau protein distribution in a 3D reconstruction of Stau immunostaining in reticulated bodies in a stage 8 egg chamber. The perspective in this 3D reconstruction is of a viewer at the posterior looking towards the anterior.
Panels O-Q are of Stau (Q and red in O) and Me31B∷GFP (P and green in O) in an egg chamber with reticulated bodies. Endogenous Stau exits reticulated bodies in stage 9 egg chambers and compacts tightly against the posterior cortex where it still colocalizes with Me31B∷GFP. Scale bars are 2.5μm in K and L, 10μm in panels B,F,J,M,N and 20μm in panels A,C,D,E, G-I and O-Q.
Published images of sponge bodies include some that correspond to the dispersed type of bodies (e.g. (Wilhelm et al., 2000), and some that appear to be a combination of dispersed and reticulated bodies (Wilsch-Brauninger et al., 1997; Theurkauf and Hazelrigg, 1998; Nakamura et al., 2001; Nakamura et al., 2004; Lin et al., 2006). It is not common to see the more extreme type of reticulated body, such as that shown in Fig. 3B, likely because the conditions required for its formation were only partially met. The presence of multiple sponge body markers in the both types of bodies makes a strong case that each represents a form of sponge body. The notion that they are extreme versions at the ends of a spectrum of a common type of structure is reinforced by the coincident reduction in the number of dispersed bodies when the reticulated bodies are present. The dispersed bodies are most similar in appearance to P bodies.
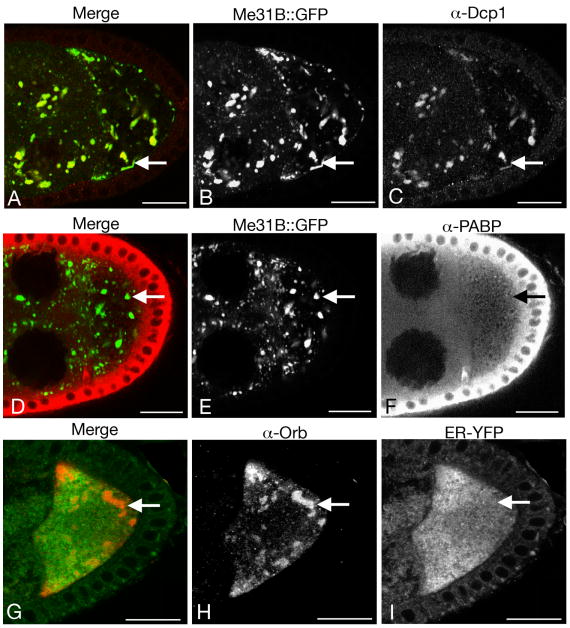
Stress granules are, like the reticulated bodies, a large type of large RNP particle, and are found in many cell types that share some components in common with P bodies (Anderson and Kedersha, 2008). To ask if the reticulated bodies are equivalent to stress granules, the distributions of poly(A) binding protein (PABP) and Dcp1 (involved in mRNA decay) were tested. PABP is present in stress granules while Dcp1 is excluded (Kedersha et al., 1999; Ingelfinger et al., 2002; Kedersha et al., 2005). We find that, conversely, Dcp1 is in the reticulated bodies (Fig. 4A-C) while PABP is not (Fig. 4D-F). By these criteria, the reticulated bodies are unlike stress granules.
Fig. 4. Reticulated bodies are not stress granules.
Each row in panels A-I shows a stage 8 egg chamber from a female maintained under conditions that promote formation of the reticulated bodies. Double labeling was used to assess protein colocalization.
A-C: Me31B∷GFP (B and green in A) and Dcp1 (C and red in A). Dcp1 is highly concentrated in the reticulated bodies.
D-F: Me31B∷GFP (E and green in D) and PABP (F and red in D). PABP is dispersed throughout the cytoplasm (with a high concentration in follicle cells, which encompass the egg chamber) and is not enriched in reticulated bodies (the arrows point to one of the bodies).
G-I: Orb (H and red in G) and ER-EYFP (I and green in G). The reticulated bodies do not exclude ER (the arrows point to one of the bodies). This egg chamber shows the more extreme type of reticulated bodies. Scale bars are 20 μm in all panels.
Sponge bodies are associated with endoplasmic reticulum (ER) (Wilsch-Brauninger et al., 1997; Wilhelm et al., 2005). Changes in ER organization could underlie the bias towards formation of the different types of sponge bodies. To test this possibility the ER marker ER-EYFP (EYFP fused to the KDEL ER retention signal; (LaJeunesse et al., 2004)) was examined in egg chambers with either dispersed bodies or reticulated bodies. In each case ER-EYFP was dispersed throughout the nurse cells and oocyte (Fig. 4G-I; data not shown). Thus, there is no detectable concentration of ER coincident with the reticulated bodies, and the shift towards formation of the reticulated bodies occurs in the absence of any obvious change in the ER.
osk mRNA localization and sponge bodies
Stau concentrates in P bodies in Drosophila neurons (Barbee et al., 2006) and in Drosophila S2 cells in culture (Eulalio et al., 2007). Therefore, given the similarities between P bodies and sponge bodies, it might be expected that Stau would also be concentrated in sponge bodies. However, published accounts of the distribution of Stau protein do not suggest such an association (St Johnston et al., 1991; Hachet and Ephrussi, 2004; Zimyanin et al., 2008). We examined the distribution of Stau under conditions when each of the extreme types of sponge bodies are present. In a stage 8 egg chamber with dispersed bodies, some of the bodies contain Stau but the bulk of Stau is spread throughout the cytoplasm in an irregular pattern (Fig. 5A,B). Conditions that induce the formation of reticulated bodies lead to a dramatic change in the distribution of Stau protein: now it is highly concentrated in the reticulated bodies (Fig. 5C). Within the reticulated bodies Stau is compartmentalized: while Me31B∷GFP levels are relatively uniform throughout the bodies, Stau is restricted to subdomains (Fig. 5L).
Stau protein plays a central role in the post-transcriptional regulation of osk mRNA, and the two molecules are normally colocalized in the oocyte (St Johnston et al., 1991). The striking difference in Stau distribution in the presence of dispersed vs. reticulated bodies raises the question of whether the distribution of osk mRNA also differs. Detection of osk mRNA by in situ hybridization reveals that it does closely parallel the distribution of Stau, independent of which type of cytoplasmic body is present (Fig. 3O,P and Fig. 5C,D). Furthermore, osk mRNA is also restricted to subdomains of the reticulated bodies, just like Stau protein (Fig. 5K).
One consequence of the formation of the reticulated bodies is the distribution of foci of osk mRNA at discrete positions within the interior of the oocyte, a pattern not previously reported in wild type ovaries. The proper localization and translational regulation of osk mRNA is critical for the subsequent abdominal patterning of the embryo. To determine if the unusual distribution of osk mRNA in reticulated bodies interferes with osk activity, we monitored development of embryos from females maintained under conditions that promote formation of the different types of sponge bodies. Cuticles of embryos from both sets of females were normal, indicating that the presence of reticulated bodies does not affect body patterning. We also monitored the fraction of eggs that hatch, a more general measure of early embryonic development. Almost all of the embryos hatched, whether they were from mothers with reticulated bodies (92% n=161), or from mothers with dispersed bodies (93% n=403).
The dramatic differences in osk mRNA distribution in egg chambers with the different types of cytoplasmic bodies raises the question of whether the mechanism of osk mRNA localization to the posterior pole of the oocyte will also be different. To begin to address this question we wished to compare the movements of osk mRNA under the different conditions. The tight colocalization of Stau protein and osk mRNA allows for indirect tracking using a Stau∷GFP fusion protein. A large fraction of Stau∷GFP faithfully mimics the distribution of endogenous Stau and is localized to the posterior of the oocyte. However, there are also large artifactual foci of Stau∷GFP whose appearance and distribution do not reflect that of endogenous Stau (Schuldt et al., 1998; Martin and St Johnston, 2003; Zimyanin et al., 2008). In our experiments we also observed the large artifactual foci, which are clearly distinct from dispersed and reticulated bodies, both of which are also labeled with Stau∷GFP. The artifactual particles were ignored in the analysis (they are noted in the figure legends).
In live egg chambers with dispersed bodies, Stau∷GFP appears in many small particles before the onset of osk mRNA localization at stage 8 (Fig. 5J). Some or all of these are presumably osk RNA transport particles. Endogenous Stau in fixed tissue has a distribution similar to that of Stau∷GFP imaged in live tissues, but is more cytoplasmic and less particulate (Fig 5B,J). The Stau∷GFP particles do match the size and distribution of osk mRNA transport particles imaged via tethering to MS2∷GFP (Zimyanin et al., 2008), suggesting that the osk RNA transport particles are not preserved with our fixation method or that tagging Stau with GFP and tethering osk mRNA to MS2-GFP increases the size of the RNA transport particles.
In live egg chambers with reticulated bodies, Stau∷GFP is present in structures identical in appearance to the reticulated bodies. The distribution of reticulated bodies containing Stau∷GFP changes during development. In early stage 8 egg chambers Stau∷GFP is associated with reticulated bodies throughout the oocyte (Fig. 5E). As stage 8 progresses, Stau∷GFP begins to concentrate in the reticulated bodies close to but not immediately contacting the posterior of the oocyte (Fig. 5G). The posterior concentration continues, with loss of Stau∷GFP from the anterior, and by stage 9 Stau∷GFP is depleted from most of the oocyte and is restricted to a narrow cortical zone at the posterior (Fig. 5H). The transition of Stau∷GFP to the posterior and out of the bulk of the oocyte does not represent a repositioning of reticulated bodies, as they can still be seen throughout the oocyte in Me31B∷GFP egg chambers of similar stages (Fig. 5O,P).
Although the final step in movement of osk transport particles to the posterior of the oocyte separates them from most of the reticulated bodies, Me31B∷GFP still colocalizes with Stau at the extreme posterior of the oocyte (Fig. 5O-Q). Similarly, other sponge body components are also enriched at this position (Wang and Hazelrigg, 1994; Wilhelm et al., 2000; Nakamura et al., 2001; Wilhelm et al., 2003; Nakamura et al., 2004; Wilhelm et al., 2005; Lin et al., 2006). Thus, the final position at which osk mRNA is localized still contains sponge body components, although they are not as highly enriched at that position as is Stau [compare P (Me31B∷GFP) and Q (Stau) in Fig. 5].
To consider possible types of Stau∷GFP movement that could account for the observed progression of posterior localization, we first obtained a three dimensional image of the reticulated bodies. The bodies were labeled for live imaging with Me31B∷GFP, and were also analyzed in fixed samples using Me31B∷GFP and Stau immunostaining. The distribution of reticulated bodies was the same using both approaches. Serial sections were reconstructed into a 3D image using Imaris software (Bitplane Inc.). A snapshot of the 3D image (Fig. 5M) and volume rendering of Stau immunostaining near the posterior (Fig. 5N) highlights the reticulated nature of these bodies. The interconnectivity shown in the reconstruction of a portion of a stage 8 oocyte (Fig. 5M) is probably an underestimate, as very thin connections between different parts of the structure may be beyond the limit of resolution by light microscopy. Thus, in stage 8 oocytes it is possible that most or all reticulated bodies in the oocyte are linked together in the more extreme examples.
The extensively interconnected nature of the reticulated bodies, whether partial or complete, allows for the possibility that the Stau/osk localization RNP complex is moved through the bodies to the posterior. In this model the reticulated bodies would serve as tracks for movement. The alternate option is that posterior directed movement occurs in the cytoplasm, and that localization RNP complexes are transiently displaced from the reticulated bodies to allow movement. The first model predicts that there should be a displacement of Stau along the reticulated bodies, while the second predicts a more uniform exchange of Stau between the reticulated bodies and the cytoplasm. These predictions can be tested in photobleaching experiments.
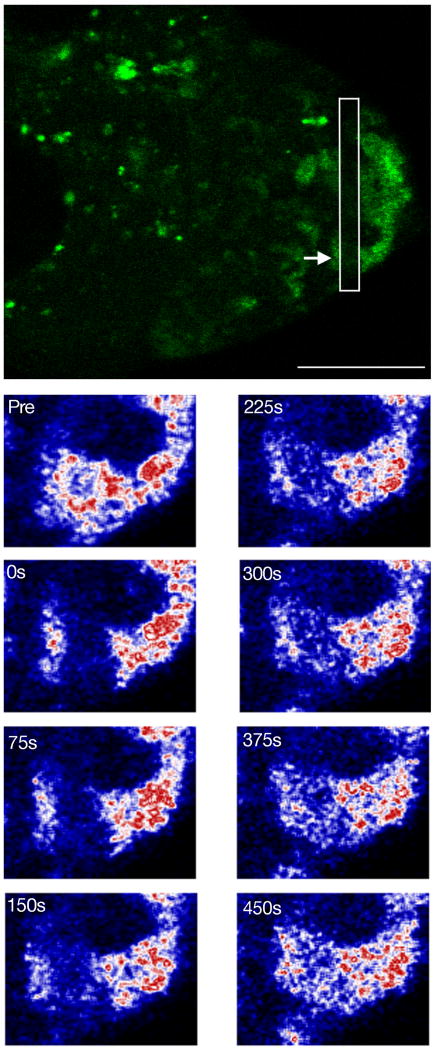
Stau∷GFP was photobleached in a band traversing an extended arm of a reticulated body that was oriented toward the posterior pole of a stage 8 oocyte (Fig. 6). For the time scale of the experiment, the laser intensity used causes effectively permanent bleaching; thus, any recovery of fluorescence is due to movement of Stau∷GFP from an unbleached zone into the bleached area. Recovery of fluorescence within the bleached band was detectable within 150 seconds, and quite substantial by 450 seconds. Notably, there was no significant displacement of the bleached band along the reticulated body. If most or all of Stau∷GFP was being moved in one direction, then we would have seen a restoration of fluorescence selectively at the more anterior boundary of the bleached band as unbleached Stau∷GFP moved towards the posterior. This was not observed. Instead, the recovery of fluorescence was relatively uniform within the bleached band, which is consistent with a random exchange of Stau∷GFP between the reticulated body and cytoplasm.
Fig. 6. Exchange of Stau between reticulated bodies and cytoplasm.
The top panel shows Stau∷GFP in a stage 8 egg chamber immediately after photobleaching. The white box surrounds the photobleached region and the arrow indicates the photobleached reticulated body shown in the bottom panels. The photobleached region is close to the posterior of the oocyte. Scale bar is 20 μm.
The lower panels are of Stau protein in a close up of a reticulated body before bleach (Pre) and at various times after photobleaching (times indicated in seconds). The oocyte posterior is to the right of the image. Stau∷GFP fluorescence is reacquired evenly throughout the photobleached region. The image of Stau∷GFP was artificially colored by converting its Look Up Table (LUT) from green to the Union Jack LUT in ImageJ (NIH). In this LUT color changes from black to blue to white to red with progressively higher fluorescence levels.
Discussion
Sponge bodies are defined as endoplasmic reticulum (ER)-like cisternae or vesicles embedded in an electron dense matrix that excludes most ribosomes (Wilsch-Brauninger et al., 1997). The first sponge body component to be identified was Exu (Wilsch-Brauninger et al., 1997), and characterization of additional components led to two generalizations. The first is that sponge bodies are enriched in post-transcriptional regulatory factors. This feature is consistent with models in which RNP particles containing regulated mRNAs are stored in sponge bodies, traffic through sponge bodies, become remodeled in sponge bodies, or a combination of all these possibilities. Our analysis extends this theme. Of the proteins we have shown to colocalize with sponge bodies, all but one have been implicated in RNA binding, gene regulation, or both. The exception is the Growl protein, which has not yet been characterized genetically and whose function is unknown.
The second generalization about sponge bodies is that they are closely related to P bodies. This is based on similarities in composition, as well as the absence or paucity of ribosomes in P and sponge bodies, respectively. Components shared by the two types of bodies include Me31B/Dhh1, Dcp1, Dcp2, eIF4E, and Tral/Car1 (Nakamura et al., 2001; Sheth and Parker, 2003; Nakamura et al., 2004; Andrei et al., 2005; Ferraiuolo et al., 2005; Wilhelm et al., 2005; Lin et al., 2006; Squirrell et al., 2006). Given the parallels between the two types of cytoplasmic bodies, it is perhaps not surprising that the terms P body and sponge body have in some cases been used synonymously. However, by structural criteria the bodies are not identical. Sponge bodies are defined as having a membranous organization (Wilsch-Brauninger et al., 1997). Ultrastructural studies of P bodies have not been reported, but the closely related GW182 bodies contain no membrane (Eystathioy et al., 2002; Yang et al., 2004; Schneider et al., 2006). What then is the relationship between these bodies? The studies reported here bear on this question, and lead us to suggest that they represent different levels of a hierarchy of cytoplasmic assemblies of RNP particles, as described below.
Several aspects of our work reveal the plasticity of sponge bodies and their dynamic organization. Previous studies of sponge bodies have focused on components that are present in both nurse cells and the oocyte (Wilsch-Brauninger et al., 1997; Nakamura et al., 2001). Our identification of Bru and Orb as sponge body components allowed for two advances. First, because these proteins are very heavily biased in their distribution between nurse cells and the oocyte – at stages 8 and 9 most Bru is in the nurse cells and almost all Orb is in the oocyte – sponge body composition could be shown to vary between the two cell types. Second, we could monitor the loss and addition of Bru and Orb following movement of sponge bodies into the oocyte. Both changes occur within close proximity to the ring canals. Given the rate of sponge body movements through ring canals and out into the ooplasm, changes in sponge body composition must occur very rapidly. Thus, the organization of sponge bodies must be dynamic. Our results are in good agreement with another study that monitored the fate of Exu∷GFP after entry into the oocyte; a partial reduction in Exu∷GFP levels within sponge bodies occurred shortly after entry and within a radius similar to what we found (Mische et al., 2007). Oocyte sponge bodies that are more distant from the ring canals have higher levels of Exu∷GFP, indicating that the loss of Exu∷GFP from sponge bodies following oocyte entry is transient.
The plasticity of sponge bodies was revealed by our discovery that their architecture is dramatically altered in different environments. Under optimal laboratory conditions sponge body components are present at readily detectable levels relatively uniformly throughout the cytoplasm, as well as in small puncta in both nurse cells and the oocyte. Under slightly less optimal conditions, but with nutrients still in abundant supply, sponge body components are concentrated in comparatively large and extensively networked reticulated bodies. The dispersed bodies and reticulated bodies are clearly related, as both contain each of the sponge body markers that were tested. Importantly, neither type of body arises as a fixation artifact, as each appears the same in live and fixed specimens. The simplest interpretation of the relationship between the two types of bodies is that they represent extremes in a spectrum of sponge body assemblies, and that different environmental conditions dictate a preference for one over the other. Variation in published descriptions of sponge bodies and their composition could well be due to relatively subtle differences in fly husbandry. Most published descriptions of sponge bodies appear to have used preparations that are not restricted to just one of the extreme types of sponge body.
Oogenesis proceeds normally even when sponge bodies are limited to just one of the two extreme types, at least to the extent of producing viable embryos that develop with no obvious morphological defects. Thus, shifting between the two types of sponge bodies appears to incur no significant liabilities.
A striking feature of the reticulated bodies is the inclusion of Stau protein and osk mRNA, which are not detectably enriched in the dispersed bodies. The progressive concentration of both molecules to the more posterior parts of the reticulated bodies raised the possibility that posterior-directed movement occurs within the bodies, which could be functioning as conduits. The evidence from photobleaching experiments ruled this out. We conclude that the microtubule dependent movements within the cytoplasm that have been implicated in localization of osk mRNA (St Johnston, 1995; Cha et al., 2002; Zimyanin et al., 2008) are likely to drive localization even when the reticulated bodies are present. A similar model has been proposed for grk RNA, which is transported in small particles and then anchored in sponge bodies at the anterior dorsal region of the oocyte (Delanoue et al., 2007).
We suggest that the different types of cytoplasmic bodies detected in the ovary represent different hierarchical assemblies of particles. By this scheme the dispersed sponge bodies would arise from assembly of smaller particles on a framework consisting of a subset of ER domains. The smaller particles would most likely be heterogeneous in nature, containing P bodies or their Drosophila equivalent as well as other types of RNP particles. This assembly is limited in that sponge body components are also present at readily detectable levels in the cytoplasm. The cytoplasmic pools of these components could represent soluble protein, or protein in submicroscopic particles. Under different environmental conditions, and for unknown reasons, the larger reticulated bodies would assemble with more complete incorporation of components and with novel components. The evidence for more complete incorporation of components is that the level of uniform cytoplasmic staining of sponge body components decreases when the reticulated bodies form. The evidence of inclusion of novel components comes from the demonstration that Stau protein and osk mRNA are present in the reticulated bodies. The compartmentalization of Stau protein and osk mRNA within the reticulated bodies argues that the assembly of these bodies is ordered, and is not simply aggregation. Compartmentalization might reflect a need to segregate mRNAs under different forms of regulation.
Experimental Procedures
Fly strains and culture
w1118 was used as wild type. The P[exu∷GFP-5] transgenic line was established using a plasmid constructed by Jim Wilhelm. Flies expressing stau∷GFP were from Daniel St Johnston (Schuldt et al., 1998; Martin et al., 2003). GFP Protein Trap lines P(PTT-GA)CG10686G89 (tralG89), P(PTT-GA)CG10686G140, and P(PTT-GA)CG14648ZCL3169 (Morin et al., 2001) were from Lynn Cooley and Bill Chia. The me31B∷GFP GFP trap line was isolated as a second insertion from the GFP trap line ZCL1796, which also had an insertion in the CG3634 gene that did not produce GFP protein. PBac(PB)growlc02107 (growlc02107) was obtained from the Exelixis collection at Harvard Medical School. The P[UAS-imp∷GFP] flies were from Cuiyun Geng. All other stocks were from the Bloomington Stock Center.
Fly food was based on the standard University of Texas recipe, which contains, for each liter of water used, 76 g corn meal, 76 ml Karo syrup, 18 g brewers yeast, 9 g agar, 1 g nipagin, 111 ml malt extract, 5 ml propionic acid and 5 ml ethanol.
Immunohistochemistry
Ovaries were fixed for 5 min and stained as described (Snee and Macdonald, 2004). The immunostaining protocol (Findley et al., 2003) preserved sponge body structures, as the distribution of Exu∷GFP was the same in live and fixed egg chambers (data not shown). Live imaging was performed as previously described previously (Snee and Macdonald, 2004).
For preparation of anti-Growl antibodies, a EcoRI/SalI fragment of the LD30155 cDNA (Genbank accession AY058608) was cloned into pET21c. This fragment encodes 502 of the 545 residues in full length Growl, and protein expressed in E. coli was used to raise rabbit polyclonal antibodies (Proteintech Group, Inc., Chicago, IL). Primary antibodies used were rabbit anti-Tral (1/500), rabbit anti-Growl (1/500), rat anti-Bru (1/500), rabbit anti-Bru (1/1000), rat anti-Stau (1/1000), rabbit anti-Hrb27C (1/2000, from Don Rio), mouse anti-Orb 4H8 (1/500, from the Developmental Studies Hybridoma Bank), rabbit anti-Dcp1 (1/20, from Tze-Bin Chou), mouse anti-PABP 6E2 (1/500, from Gideon Dreyfuss). Each sponge body associated protein displayed the same distribution independent of the presence of the GFP sponge body markers.
In situ hybridization
Fluorescence in situ hybridizations to whole mount ovaries were performed by fixing the ovaries with 4% formaldehyde in PBS for 20 minutes, washing in PBST (PBS plus 0.1% Triton X-100), then processing for pre-hybridization, hybridization and post-hybridization washes as described (Tautz and Pfeifle, 1989), using antisense probes labeled with digoxigenin, and previously described modifications (Kim-Ha et al., 1991). After the post-hybridization washes the samples were incubated with anti-digoxigenin-HRP (diluted 1/200; Roche) for 1-2 hours, washed multiple times in PBST and incubated in Tyramide-Cy5 reagent (diluted 1/50; Perkin Elmer) in the manufacturers amplification diluent for 30 minutes, washed in PBST several times, and mounted on slides (Hughes and Krause, 1999; Wilkie et al., 1999; Vanzo and Ephrussi, 2002).
Immunoprecipitation
Immunoprecipitations were carried out as described (Wilhelm et al., 2000) using fresh ovarian extract and DXB150 homogenization buffer (25 mM Hepes, pH 6.8, 150 mM KCl, 1 mM MgCl2, 1 mM DTT, 250 mM sucrose, and 1% Triton X-100).
Supplementary Material
Panels A and B show Growl immunostaining in wild type (A) and growlc02107/growlc02107 (B) egg chambers. Growl protein is expressed in germ line cells throughout oogenesis and is maternally deposited into embryos (not shown). The Growl immunostaining (A) is specific for this protein as it is absent in egg chambers from growl mutant mothers (B) that lack Growl protein (see below). The low staining in the central regions of later stage egg chambers in A is typical of the pattern when antibody penetration is poor. Scale bars are 20 μm. Panel C shows western blot detection of Growl and b-tubulin in extracts from w1118 (wt) and growlc02107/Df(3R)ED5046 (growl-) ovarian extracts. The growlc02107 allele appears to be a protein null allele.
Acknowledgments
We thank Daniel St Johnston, Lynn Cooley, Bill Chia, Cuiyun Geng and the Bloomington stock center for fly stocks, and Bill Chia for generating the g89 and g140 GFP trap lines. Paul Lasko, Mary Lilly, Tze-Bin Chou, Gideon Dreyfuss and Don Rio generously provided antibodies. We thank the Developmental Studies Hybridoma Bank at The University of Iowa for the monoclonal antibody 4H8 (P. Schedl), and the Berkley Drosophila Genome Project for the plasmids containing the growl and Pcm cDNAs. We thank Angela Bardo for Imaris software training and Christian Antonio for cloning the CG14648 expression construct. This work was supported by grants GM42612 and GM54409 from the NIH.
Grant Sponsor: National Institutes of Health; Grant numbers GM42612 and GM54409
References
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook. Vol. 434 1989. [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Ann Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Boylan KL, Mische S, Li M, Marques G, Morin X, Chia W, Hays TS. Motility screen identifies Drosophila IGF-II mRNA-binding protein--zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet. 2008;4:e36. doi: 10.1371/journal.pgen.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha BJ, Serbus LR, Koppetsch BS, Theurkauf WE. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat Cell Biol. 2002;4:592–598. doi: 10.1038/ncb832. [DOI] [PubMed] [Google Scholar]
- Colegrove-Otero L, Minshall N, Standart N, Wickens M. RNA-Binding Proteins in Early Development. Crit Rev Biochem Mol Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13:523–538. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9(10):1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Geng C, Macdonald PM. Imp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol Cell Biol. 2006;26:9508–9516. doi: 10.1128/MCB.01136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Hughes SC, Krause HM. Single and double FISH protocols for Drosophila. Methods Mol Biol. 1999;122:93–101. doi: 10.1385/1-59259-722-x:93. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, Cooley L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004;32:D418–20. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila ooctye. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- King RC. Ovarian development in Drosophila melanogaster. 1970. [Google Scholar]
- LaJeunesse DR, Buckner SM, Lake J, Na C, Pirt A, Fromson K. Three new Drosophila markers of intracellular membranes. BioTechniques. 2004;36:784–8. 790. doi: 10.2144/04365ST01. [DOI] [PubMed] [Google Scholar]
- Lin MD, Fan SJ, Hsu WS, Chou TB. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell. 2006;10:601–613. doi: 10.1016/j.devcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo R, Zhou X, Shillinglaw W, Henzel W, Macdonald PM. BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol Cell Biol. 2001;21:3462–3471. doi: 10.1128/MCB.21.10.3462-3471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Leclerc V, Smith-Litiere K, St Johnston D. The identification of novel genes required for Drosophila anteroposterior axis formation in a germline clone screen using GFP-Staufen. Development. 2003;130:4201–4215. doi: 10.1242/dev.00630. [DOI] [PubMed] [Google Scholar]
- Martin SG, St Johnston D. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature. 2003;421:379–384. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- Mische S, Li M, Serr M, Hays TS. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell. 2007;18:2254–2263. doi: 10.1091/mbc.E06-10-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro TP, Kwon S, Schnapp BJ, St Johnston D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J Cell Biol. 2006;172:577–588. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila Cup Is an eIF4E Binding Protein that Associates with Bruno and Regulates oskar mRNA Translation in Oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Cilius Nielsen F, Kragh Jakobsen R, Christiansen J. The biphasic expression of IMP/Vg1-RBP is conserved between vertebrates and Drosophila. Mech Dev. 2000;96:129–132. doi: 10.1016/s0925-4773(00)00383-x. [DOI] [PubMed] [Google Scholar]
- Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldt AJ, Adams JHJ, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes & Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Bicaudal C and Trailer hitch have similar roles in gurken mRNA localization and cytoskeletal organization. Developmental Biol. 2009 doi: 10.1016/j.ydbio.2009.02.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. mRNA 5′ Cap-binding Protein elF4E and Control of Cell Growth. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1996. pp. 245–269. [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster(1) Cold Spring Harbor Laboratory Press; New York: 1993. pp. 1–70. [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development. 1989;107:13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals a translational control of the segementation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hazelrigg TI. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development. 1998;125:3655–3666. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo NF, Ephrussi A. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development. 2002;129:3705–3714. doi: 10.1242/dev.129.15.3705. [DOI] [PubMed] [Google Scholar]
- Wang S, Hazelrigg T. Implications for bcd mRNA localization from spatial distribution of exu protein in Drosophila oogenesis. Nature. 1994;369:400–403. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD. Isolation of a Ribonucleoprotein Complex Involved in mRNA Localization in Drosophila Oocytes. J Cell Biol. 2000;148:427–440. doi: 10.1083/jcb.148.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Shermoen AW, O'Farrell PH, Davis I. Transcribed genes are localized according to chromosomal position within polarized Drosophila embryonic nuclei. Curr Biol. 1999;9:1263–1266. doi: 10.1016/s0960-9822(99)80509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsch-Brauninger M, Schwarz H, Nusslein-Volhard C. A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol. 1997;139:817–829. doi: 10.1083/jcb.139.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panels A and B show Growl immunostaining in wild type (A) and growlc02107/growlc02107 (B) egg chambers. Growl protein is expressed in germ line cells throughout oogenesis and is maternally deposited into embryos (not shown). The Growl immunostaining (A) is specific for this protein as it is absent in egg chambers from growl mutant mothers (B) that lack Growl protein (see below). The low staining in the central regions of later stage egg chambers in A is typical of the pattern when antibody penetration is poor. Scale bars are 20 μm. Panel C shows western blot detection of Growl and b-tubulin in extracts from w1118 (wt) and growlc02107/Df(3R)ED5046 (growl-) ovarian extracts. The growlc02107 allele appears to be a protein null allele.