Abstract
The phytopathogenic basidiomycetous fungus Ustilago maydis secretes, under conditions of nitrogen starvation, large amounts of the biosurfactant ustilagic acid (UA). This secreted cellobiose glycolipid is toxic for many microorganisms and confers biocontrol activity to U. maydis. Recently, a large gene cluster that is responsible for UA biosynthesis was identified. Here, we show that expression of all cluster genes depends on Rua1, a nuclear protein of the C2H2 zinc finger family, whose gene is located within the gene cluster. While deletion of rua1 results in complete loss of UA production, overexpression of rua1 promotes increased UA synthesis even in the presence of a good nitrogen source. Bioinformatic analysis allowed us to identify a conserved sequence element that is present in the promoters of all structural genes involved in UA biosynthesis. Deletion analysis of several promoters within the cluster revealed that this DNA element serves as an upstream activating sequence (UAS) and mediates Rua1-dependent expression. We used the yeast one-hybrid system to demonstrate specific recognition of this DNA element by Rua1. Introduction of nucleotide exchanges into the consensus sequence interfered with Rua1-dependent activation, suggesting that this sequence element acts as a direct binding site for Rua1.
Many fungi have the ability to synthesize complex secondary metabolites (15). These bioactive compounds are not essential for viability and are often produced only under certain environmental conditions or at selected times during the life cycle of the organism. Many secondary metabolites help the fungal cells to cope with environmental stress or to compete with other microorganisms.
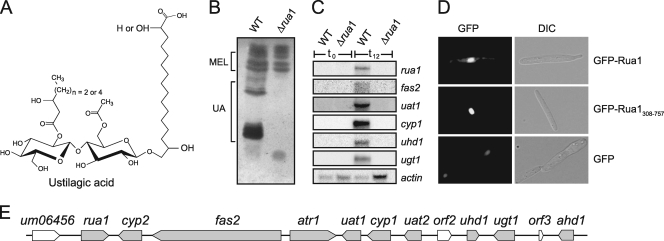
Under conditions of nitrogen limitation, the basidiomycete Ustilago maydis produces large amounts of extracellular glycolipids (4). Glycolipids are amphipathic substances and thus act as biosurfactants. Some of the known extracellular glycolipids also display antibiotic activity. U. maydis is unique among fungal producers of glycolipids since it secretes two structurally unrelated classes of glycolipids. The mannosylerythritol lipids (MELs) are heavier than water and form an extracellular oil (9). MELs are highly potent surface-active substances but not toxic for other organisms (19). In addition, U. maydis produces the cellobiose lipid ustilagic acid (UA) (see Fig. 2A), which has broad antibacterial and antifungal spectra (11). The enzymes involved in biosynthesis of these glycolipids are in both cases encoded by genes found at adjacent locations on the chromosome. While a small gene cluster of five genes on chromosome 7 is responsible for MEL production, a large gene cluster on chromosome 23, encompassing 45 kb and containing 12 open reading frames (ORFs), is involved in UA production (13, 29). The UA gene cluster contains all genes necessary for the production of this glycolipid, including genes encoding two cytochrome P450 monooxygenases (cyp1 and cyp2), an acyl- and an acetyl-transferase (uat1 and uat2), a complete single-chain fatty acid synthase (fas2), a glycosyl transferase (ugt1), two hydroxylases (uhd1 and ahd1), and a potential export protein of the ABC-transporter family (atr1) (see Fig. 2E). By mutational analysis and mass spectrometric analysis of secreted intermediates, we were able to deduce the complete biosynthesis pathway of the cellobiose lipid UA (29).
FIG. 2.
Rua1 is essential for UA biosynthesis. (A) Molecular structure of ustilagic acid (UA). UA consists of cellobiose O-glycosidically linked to the ω-hydroxyl group of 15,16-dihydroxypalmitic acid or 2,15,16-trihydroxypalmitic acid. The cellobiose moiety is esterified with an acetyl group and a short-chain β-hydroxy fatty acid. (B) TLC analysis of extracellular glycolipids produced by wild-type and Δrua1 mutant strains. Deletion of rua1 results in complete loss of UA secretion, while production of MELs is not affected. (C) Total RNA of wild-type and rua1 deletion strains was prepared before (0 h) and 12 h after shifting of cells to nitrogen-limited medium. Under these conditions, transcription of cluster genes is strongly induced in wild-type strains but is completely abolished in rua1 mutant strains. The actin gene is constitutively expressed and serves as a loading control. (D) Rua1 localizes to the nucleus. U. maydis FB2 strains expressing full-length eGFP-Rua1 or the N-terminally truncated version eGFP-Rua1308-757 were analyzed by differential interference contrast (DIC) light microscopy (right column) and epifluorescence (left column) to detect eGFP. Both fusion proteins show strong localization to the nucleus. Expression of the unfused eGFP reporter gene serves as a control (GFP). (E) Genetic organization of the UA biosynthesis gene cluster. For gene designations, see reference 29.
In fungi, genes involved in secondary metabolism are often arranged in clusters which are coregulated (15). In some cases, the transcription factor responsible for coregulation is itself encoded by one of the cluster genes, e.g., the Aspergillus flavus transcription factor AflR, which regulates aflatoxin and sterigmatocystin biosynthesis (35), and the regulator of gliotoxin production GliZ in Aspergillus fumigatus (3).
Here, we show that the zinc finger protein Rua1, whose gene is part of the UA biosynthesis gene cluster, acts as a transcription factor and regulates the activity of all structural genes involved in UA biosynthesis. We show that Rua1 binds directly to a short conserved DNA sequence motif, which is present in the promoters of all cluster genes and which confers nitrogen-dependent expression of UA biosynthesis.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Escherichia coli strain DH5α and Saccharomyces cerevisiae strain DF5 his1-1 (8) were used for all DNA manipulations. Construction of plasmids was performed using standard procedures (24). U. maydis strains FB1 (a1 b1), FB2 (a2 b2) (2), and MB215 (a2 b13) (12) were grown as described before (21). The U. maydis Δrua1 deletion strain and the strains in which rua1 is expressed under the control of either the constitutive Potef promoter (MB215-Potef::rua1) or the arabinose-inducible Pcrg promoter (MB215-Pcrg::rua1) were generated according to the published protocol for PCR-based generation of gene replacement mutants (5, 17). The sequences of the primers used for generation of mutants are available from the authors on request. All mutants were confirmed by Southern analysis, and deletion phenotypes were verified by complementation. Transformation of U. maydis was performed as described previously (26). For selection of transformants, PD plates containing 200 mg/ml hygromycin or 2 μg/ml carboxin were used.
To induce glycolipid production, strains were grown at 30°C in liquid yeast extract-peptone-sucrose (YEPS) medium to logarithmic phase and then shifted to nitrogen starvation medium containing 1.7 g/liter yeast nitrogen base (YNB; Difco) and 5% glucose as a carbon source. Glycolipids were isolated after cultivation of cells for 4 days at 30°C on a rotary shaker. The Potef::rua1 strain was grown in liquid YEPS medium to logarithmic phase prior to glycolipid extraction.
Localization of Rua1.
For construction of U. maydis strains expressing green fluorescent protein (GFP)-Rua1, the rua1 gene and shortened versions of rua1 were amplified and cloned downstream of the GFP gene in plasmid p123, which carries an enhanced GFP (eGFP) reporter under the control of the strong constitutive Potef promoter (28). Prior to transformation, plasmids were linearized with SspI. Integration into the ip locus of U. maydis was verified by Southern blot analysis.
Northern analysis.
Total RNA was isolated for Northern blot analysis (25). Wild-type (WT) strain MB215 and the rua1 deletion strain were grown at 30°C to logarithmic phase in YNB medium containing 5% glucose and 0.2% ammonium sulfate. After centrifugation, cells were resuspended in the same volume of fresh medium containing 5% glucose but lacking a nitrogen source. RNA was prepared at the time points indicated. Northern blots were prepared using standard procedures (24). For hybridization, internal probes of the respective genes that have been amplified by PCR were used.
Isolation of glycolipids.
Extracellular glycolipids were extracted from suspension cultures (0.5 ml) with 1 volume of ethyl acetate. The ethyl acetate phase was evaporated, and glycolipids were dissolved in methanol. Glycolipids were analyzed by thin-layer chromatography (TLC) on silica plates (Silica gel 60; Merck) with a solvent system consisting of chloroform-methanol-water (65:25:4, vol/vol). The plate was dried thoroughly, and sugar-containing compounds were visualized by spraying them with a mixture of glacial acetic acid-sulfuric acid-p-anisaldehyde (50:1:0.5, vol/vol) and heating them at 150°C for 3 min (12).
Generation of truncated promoter constructs.
For the construction of the plasmids containing the different promoter fragments, DNA sequences upstream of the atr1, cyp1, and cyp2 genes were amplified by PCR and inserted into the plasmid p123-mCherry, containing the mCherry gene as a reporter gene (27, 28), thereby replacing the constitutive Potef promoter. All plasmids contained a carboxin resistance (ip) cassette as a selectable marker in U. maydis. For integrative transformation into the ip locus of U. maydis MB215-Pcrg::rua1 strains, plasmids were linearized with SspI.
Yeast one-hybrid system.
S. cerevisiae strain YM4271 (32, 33) was used for yeast one-hybrid analysis. Cells were grown in liquid yeast extract-peptone-dextrose (YEPD) medium or minimal medium lacking the appropriate uracil or leucine amino acids at 30°C to logarithmic phase. To generate the reporter plasmid, DNA fragments from the atr1 promoter were amplified by PCR and inserted into the XhoI or HindIII sites of pLacZi (Clontech, Mountain View, CA). Plasmids pAD2-rua1, pAD2-rua1NΔ1-307, and pAD2-rua1NΔ1-576 were created as follows. The rua1 gene and the N-terminally shortened versions of rua1 were amplified by PCR and cloned into pAD2 as described previously (16). Cells were cotransformed as follows. Harvested cells were resuspended in 100 μl LiT (10 mM Tris-Cl, pH 7.4, 100 mM lithium acetate) and transformed with either 1 μg of the corresponding plasmid DNA or 5 μl salmon sperm DNA (10 mg/ml). Cells were covered with 500 μl LiT-polyethylene glycol (PEG) (LiT with 1 g/ml PEG 3350) and slowly rotated for 1 h. The mixture was supplemented with 50 μl dimethyl sulfoxide (DMSO) and treated with heat shock at 42°C for 15 min. Cells were plated on YEPD medium lacking the appropriate amino acids.
For quantitative measurements, β-galactosidase activity was determined using early logarithmic-phase cultures (optical density at 600 nm [OD600] of ∼0.5) of yeast cells transformed with the indicated plasmids. Samples (3 ml) were withdrawn from the cultures, and cells were harvested, washed with 1 ml Z buffer (100 mM Na2HPO4-NaH2PO4, pH 7.0, 10 mM KCl, 2 mM MgSO4), and resuspended in 250 μl of Z buffer. The cells (100 μl) were permeabilized by three cycles of freezing and thawing. To each sample, 700 μl Z buffer, 20 μl β-mercaptoethanol, and 160 μl ONPG (o-nitrophenyl-β-d-galactopyranoside; 4 mg/ml in Z buffer) were added. Following a 10-min incubation, 400 μl of 1 M Na2CO3 was added to stop the reactions. After clarification by centrifugation, the A420 was measured, and β-galactosidase activity was calculated as Miller units (23).
Microscopy.
For microscopic analysis, logarithmically growing cells were placed on agarose cushions and directly analyzed using a Zeiss Axiovert 200 microscope (Göttingen, Germany). Images were taken using a cooled charge-coupled-device (CCD) camera (Hamamatsu Orca-ER, Bridgewater, NJ) with an exposure time of 100 to 1.000 ms.
RESULTS
The rua1 gene contains a cysteine-rich region distantly related to the classic C2H2 zinc finger motif.
The UA biosynthesis gene cluster contains a gene that encodes a protein carrying a characteristic cysteine-rich region at its C-terminal end. A BLAST search revealed that this domain displays some similarity to zinc finger transcription factors of the C2H2 family (Fig. 1). This suggested that this protein might be involved in regulation of UA biosynthesis. Therefore, this gene has been termed rua1 (regulator of ustilagic acid production) (29).
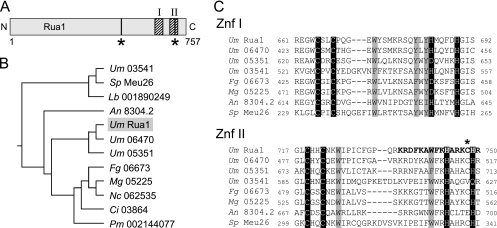
FIG. 1.
Domain organization of the transcription factor Rua1. (A) Scheme of the domain organization of Rua1. The two C-terminal zinc finger domains are indicated by hatched areas. Predicted nuclear localization motifs are marked by vertical bars and asterisks. (B) Database comparison of the C-terminal region reveals that Rua1 is related to a yet uncharacterized subfamily of fungal proteins. The tree was constructed with CLUSTALW. The GenBank accession numbers are as follows: for U. maydis (Um) 03541, XP_759688; for Schizosaccharomyces pombe (Sp) Meu26, BAB60877; for Lacaria bicolor (Lb) 001890249, XP_001890249; for A. nidulans (An) 8304.4, CBF80270; for U. maydis (Um) Rua1, XP_762605; for U. maydis (Um) 06470, XP_762617; for U. maydis (Um) 05351, XP_761498; for Fusarium gramineum (Fg) 06673, XP_386849; for Magnaporthe grisea (Mg) 05225, XP_359552; for Neurospora crassa (Nc) 062535, XP_963086; for Coccidioides immitis (Ci) 03864, XP_001244423; and for Penicillium marneffei (Pm) 002144077, XP_002144077. (C) Sequence alignment of both Rua1 zinc finger domains (ZnF I and ZnF II). The Cys and His residues predicted to be involved in coordination of zinc are shaded in black. Highly conserved amino acids are shaded in gray. The C-terminal nuclear localization motif overlaps with the Znf II sequence (bold letters); the amino acid exchanged in the rua1C747R mutant strain is marked with an asterisk.
The deduced polypeptide sequence of rua1 encodes a protein of 757 amino acid residues, which contains two potential C-terminal zinc finger domains (Fig. 1A). Database comparison revealed that Rua1 belongs to a small family of fungal proteins characterized by two putative zinc finger domains (Fig. 1B). Members of this subfamily were found both in ascomycetes and in basidiomycetes, with U. maydis containing a total of four related proteins. Two of them, Um06470 and Um05351 (for the MUMDB entry number, see http://mips.gsf.de/genre/proj/ustilago), are of unknown function but are very closely related to Rua1. A third U. maydis protein (Um03541) is similar to Schizosaccharomyces pombe Meu26, which was isolated in a screen for meiotically upregulated genes (31) (Fig. 1B).
We termed the two putative zinc finger domains Znf I and Znf II. In Rua1, they are located in the C-terminal part of the protein, comprising the positions between amino acids 663 to 690 and 718 to 749, respectively. Interestingly, the amino acid sequence of the zinc finger domains resembles that of C2H2 zinc fingers but at the same time deviates significantly from the classic C2H2 motif (34). Therefore, the Rua1 zinc finger was not predicted if database searches such as those using Interpro, SMART, or Pfam were performed. It might be surprising that for some members of the Rua1 family of zinc finger proteins, one of the histidines is exchanged for an aspartic acid. However, recently, it has been shown for type 2 B-box domain proteins that aspartic acid might also be involved in zinc atom coordination (22). Thus, Rua1 and the other related fungal proteins constitute a novel subfamily of C2H2-like zinc finger transcription factors (Fig. 1).
Rua1 is necessary for regulation of the UA biosynthesis gene cluster.
To address the physiological function of Rua1 and its involvement in cluster regulation, we constructed a deletion mutant for rua1 by replacing the open reading frame with a hygromycin resistance cassette. Wild-type and rua1 deletion strains were cultivated for 4 days under nitrogen limitation and tested for glycolipid production by thin-layer chromatography (TLC). Whereas wild-type cells synthesized large amounts of both MEL and UA glycolipids, production of UA was completely abolished in rua1 deletion strains. Secretion of MEL glycolipids, however, was unaffected in rua1 mutants (Fig. 2B).
To determine whether deletion of rua1 influences the transcriptional level of the cluster genes, Northern blot analyses were performed. In wild-type strains, all UA biosynthesis genes are strongly upregulated under conditions of nitrogen starvation (29). Six representative genes of the UA biosynthesis gene cluster were tested for expression in rua1 deletion strains. Upon nitrogen starvation, no expression of these genes could be observed (Fig. 2C). This supports the notion that Rua1 plays a major role in regulation of the UA biosynthesis cluster genes.
The program PSORT (http://psort.ims.u-tokyo.ac.jp/form.html) predicts two putative nuclear localization motifs, which are indicated in Fig. 1A. The amino acid sequences of these motifs are R468KHKVDAVRQSAVRSRR484 and R735KRDFKAWFKHARKCHR750. To verify the predicted nuclear localization of Rua1, different GFP-Rua1 fusion proteins were expressed under the control of the constitutively active Potef promoter. All these constructs were introduced into a rua1 deletion strain and analyzed for functionality. While a C-terminal full-length Rua1-GFP fusion was not functional, the respective N-terminal GFP fusion was able to trigger normal amounts of UA production in the Δrua1 strain (data not shown). If the first 307 N-terminal amino acids of Rua1 were removed, no induction of UA production was observed (data not shown). If either the full-length N-terminal GFP-Rua1 fusion protein or the truncated version (GFP-Rua1308-757) was expressed in U. maydis cells, nuclear localization was observed, indicating that Rua1 is targeted to the nucleus (Fig. 2D), most presumably due to its C-terminal nuclear localization domains. Together, this suggests that the N-terminal region of Rua1 is important for the function of Rua1 but not involved in nuclear localization.
Expression of rua1 is sufficient to support UA biosynthesis under noninducing conditions.
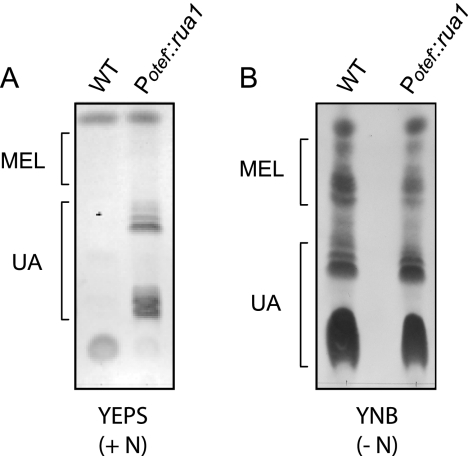
To test whether expression of Rua1 is also sufficient to trigger gene regulation of the UA biosynthesis genes, we expressed the ORF of rua1 under the control of the constitutive Potef promoter (Potef::rua1) in a strain deleted for the endogenous copy of rua1. Glycolipid production in these strains was analyzed by TLC. Cells were grown in either nitrogen-free minimal medium (YNB −N) or complete medium (YEPS). Secreted glycolipids were extracted from the culture with ethyl acetate and analyzed by TLC. Remarkably, expression of rua1 under the control of the constitutive Potef promoter resulted in UA production even in noninducing rich medium (Fig. 3A). One has to point out that MEL production was not observed in this strain under these conditions. Nitrogen depletion induced production of comparable amounts of UA both in the WT and in the strain constitutively expressing Rua1 (Fig. 3B). Taken together, these data indicate that Rua1 is both necessary and sufficient to trigger UA biosynthesis. Since production of MEL glycolipids was not affected in Rua1-expressing strains, Rua1 appears to act as a cluster-specific regulator of UA biosynthesis.
FIG. 3.
Constitutive expression of rua1 results in UA production. (A) U. maydis MB215 (WT) and MB215 Δrua1 Potef::rua1 (Potef::rua1) cells were grown in rich medium (YEPS) containing nitrogen (+ N). While wild-type cells grown with sufficient nitrogen were unable to secrete glycolipids under these conditions, constitutive expression of rua1 resulted in significant UA secretion. (B) The same strains were grown in minimal medium (YNB) lacking nitrogen (− N).
Regulation of Rua1 expression.
To elucidate the regulation of Rua1, we also analyzed the transcription of rua1 by Northern blot analysis. While no expression of rua1 could be observed if a rich nitrogen source was offered, nitrogen limitation resulted in strong rua1 expression (Fig. 2C). These Northern data thus confirm our previous results observed with microarray analysis (29).
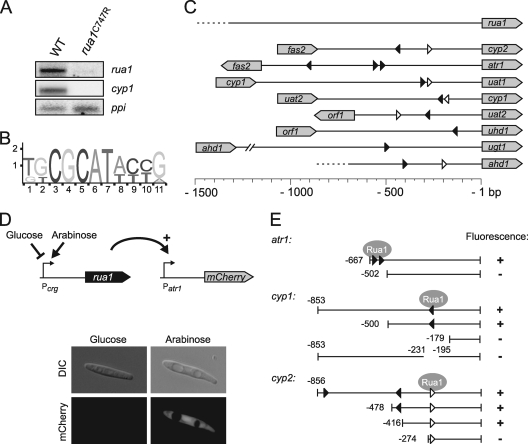
During construction of a deletion mutant for the gene cyp2, which is adjacent to rua1 and codes for a P450 monooxygenase involved in subterminal hydroxylation of hexadecanoic acid (29), we inadvertently introduced a point mutation into the coding region of rua1. This mutation affects a conserved cysteine at position 747 between the two core histidines of the second C2H2-like zinc finger (Fig. 1C, marked with an asterisk). Interestingly, this mutant was unable to produce UAs, indicating that the function of rua1 was affected by this point mutation. Northern blot analysis revealed that the nitrogen-dependent transcription of the cluster genes was abolished in the mutant strain, which expresses rua1C747R (Fig. 4A and data not shown). Interestingly, expression of rua1C747R itself was also abolished in the mutant strain, suggesting that transcription of rua1 may be subject to positive feedback regulation (Fig. 4A).
FIG. 4.
Autoregulation of Rua1 and localization of the Rua1 DNA binding motif. (A) Total RNA of wild-type MB215 and rua1 mutation strain MB215 rua1C747R was prepared 12 h after shifting of cells to nitrogen-limited medium. While wild-type cells show strong expression of the UA biosynthesis cluster genes, indicated by cyp1 expression, rua1C747R cells were not able to express cyp1. Also, the rua1 gene was not expressed in this mutant strain, indicating autoregulation of Rua1. The ppi gene is constitutively expressed and serves as a loading control. (B) A common motif within the promoter regions of the UA cluster genes was identified by MEME analysis. (C) Localization of the predicted motif within the 5′ untranslated regions of the UA biosynthesis genes. The triangles indicate the direction of the motif; black triangles indicate that the sequence matches with the predicted motif; white triangles allow for a single mismatch within the variable region. (D) Schematic diagram of the constructs used to narrow down the core region for Rua1-dependent promoters. Different promoter regions were fused to the mCherry reporter gene and transformed into a U. maydis strain expressing Rua1 under the control of the inducible Pcrg promoter. U. maydis strains expressing the mCherry gene were analyzed by DIC light microscopy (upper column) and epifluorescence (lower column). (E) Schematic diagram of the various promoter deletion constructs and their activities. The numbers indicate the lengths of the selected 5′ untranslated regions of the respective genes or refer to deleted regions. mCherry gene activity was determined by microscopy.
Promoters of Rua1-regulated genes share a conserved DNA sequence element.
To identify potential regulatory sequences within the promoter regions of the coregulated cluster genes, an in silico search for conserved sequence motifs was performed using the MEME algorithm (http://meme.sdsc.edu/meme4_1/cgi-bin/meme.cgi). This analysis revealed a short consensus sequence ([T/G][G/T]CGCAT[A/T][C/T][C/T][G/A]) (Fig. 4B) which can be found in either orientation in the promoter regions of all cluster genes in at least one copy. Interestingly, the rua1 promoter itself does not contain this element (Fig. 4C).
To determine whether the predicted sequence elements contribute to the regulation of the cluster genes, we performed a deletion analysis. The putative promoter regions used were either full-length or truncated at their 5′ ends to drive expression of the mCherry reporter gene. All constructs were integrated as single copies into the ip locus of the U. maydis strain MB215-Pcrg::rua1 by homologous recombination. In this strain, rua1 is expressed under the control of the arabinose-inducible Pcrg promoter (Fig. 4D). Strains were first grown in glucose medium and then transferred to liquid medium containing arabinose as the sole carbon source to induce rua1 expression. Expression of the mCherry gene was followed by fluorescence microscopy to determine which of the truncated promoters could still be induced by Rua1 (Fig. 4D).
A truncated version of the atr1 promoter including the region comprising bp −667 to bp −1 still showed full promoter activity while a fragment shortened to bp −502 showed no activity. This indicates that for atr1, the region between bp −667 and −502 is critical for regulation by Rua1. Interestingly, this region contains two copies of the predicted motif (Fig. 4E). Next, we selected the 853-bp intergenic region between cyp1 and uat2 for a detailed promoter analysis. The whole 853-bp 5′ untranslated sequence as well as the 500-bp shortened region conferred high mCherry fluorescence upon Rua1 induction, whereas no fluorescence was detected when the promoter region was shortened to 179 bp. If the region between positions −231 and −195, which contains the predicted motif, was deleted, promoter activity was completely abolished, supporting our view that this motif acts as an upstream activating sequence (UAS) which is critical for induction by Rua1 (Fig. 4E).
In the cyp2 promoter, the sequence from bp −478 to bp −1 was sufficient to confer Rua1 dependent expression to the same extent as the entire intergenic region. Surprisingly, Rua1 regulation was still observed if the promoter was shortened to 416 bp and thus did not contain a consensus motif predicted within this region. However, a more degenerate sequence motif containing a nonconserved nucleotide within the flanking area is located further downstream. Therefore, we tested if the promoter region comprising bp −274 to −1, which contains this motif at the very left border, still supports Rua1-dependent mCherry gene expression. Surprisingly, no activity was induced, suggesting that the function of the consensus motif may in this case depend on its immediate sequence context, which was altered in this reporter construct (Fig. 4E).
Rua1 specifically recognizes the conserved upstream activating sequence present in the promoters of all UA biosynthetic cluster genes.
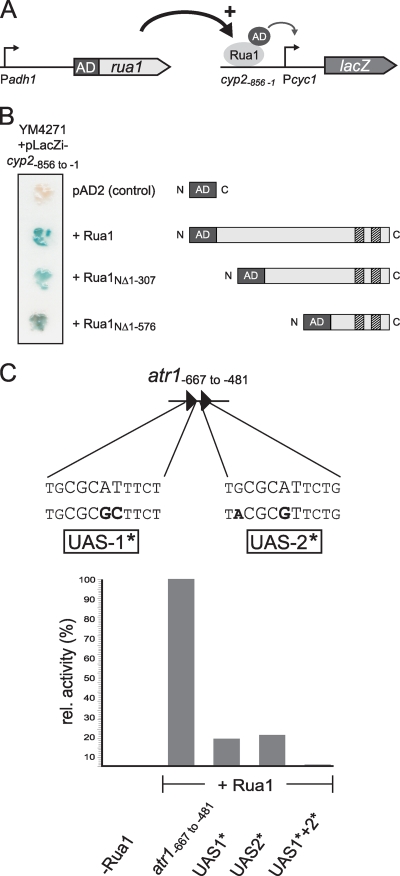
To verify that Rua1 acts as a transcriptional factor and is capable of binding DNA, we tried to perform in vitro DNA binding assays. Unfortunately, expression of Rua1 in E. coli resulted in the formation of inclusion bodies that could not be solubilized. Therefore, we used the yeast one-hybrid system to test whether the conserved sequence motif is recognized by Rua1 in vivo. To this end, the activation domain of the yeast Gal4 protein was fused either to full-length Rua1 or to N-terminally truncated versions (Rua1NΔ1-307 and Rua1NΔ1-576). A plasmid expressing only the Gal4 activation domain served as a control. Plasmids were transformed into S. cerevisiae strain YM4271, which harbors a reporter construct in which the regulatory region of the cyp2 promoter was placed upstream of the minimal promoter of the iso-1-cytochrome c (CYC1) gene, which itself drives the reporter gene lacZ (Fig. 5A). The activity of the chimeric promoters was quantified by growing S. cerevisiae on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) containing solid medium. Blue coloring of colonies indicated β-galactosidase activity. The results shown in Fig. 5A demonstrate that full-length Rua1 was clearly able to activate β-galactosidase activity. This indicates that Rua1 expression is sufficient to stimulate expression of the chimeric promoter in yeast. Furthermore, the N-terminally shortened versions of Rua1 were also able to activate reporter gene activity, indicating that the C-terminal zinc finger domain of Rua1 is sufficient to recognize the putative upstream activating sequence. In contrast, control strains which express only the Gal4 activation domain showed no β-galactosidase activity.
FIG. 5.
Rua1 binds to a conserved DNA motif within the promoter region of the UA cluster genes. (A) Schematic diagram of constructs used in the yeast one-hybrid system. A chimeric protein where Rua1 or N-terminally shortened versions Rua1NΔ1-307 and Rua1NΔ1-576 were N-terminally fused to the activation domain of Gal4 was constructed and expressed under the constitutive yeast alcohol dehydrogenase promoter (Pahd1). As a reporter construct, the 5′ untranslated region of the cyp2 gene (Pcyp2) containing the Rua1 binding site was introduced upstream of the minimal promoter of the iso-1-cytochrome c (Pcyc1) gene, which was fused to the reporter gene lacZ. (B) Yeast one-hybrid transcription activation analysis of Rua1 or N-terminally shortened versions of Rua1. S. cerevisiae strain YM4271 was cotransformed with the Gal4 activation domain alone or Gal4-AD-Rua1 fusion proteins and the reporter plasmid. Transcription activation was visualized by adding X-Gal which was cleaved by β-galactosidase, yielding an insoluble blue product. (C) Schematic diagram of the construct used in the yeast one-hybrid system. An ∼200-bp fragment of the atr1 promoter (atr1−667 to −481), containing two times the motif identified by MEME analysis, was used for Rua1 binding studies. The motifs found in this area, proposed to act as upstream activating sequences (UAS-1 and UAS-2) for Rua1, are indicated. Black letters indicate nucleotides that have been mutated in this study to give UAS-1* and UAS-2*. For quantification, β-galactosidase activity was determined as described in Materials and Methods and is indicated in Miller units. Shown are relative activities compared to the activity of the 200-bp atr1 UAS, which was set to 100%.
To further prove that Rua1 directly binds to the predicted sequence motif, we tested whether mutation of conserved residues within the consensus sequence interferes with Rua1 activation. To this end, a fragment of the upstream region of atr1 was analyzed by yeast one-hybrid experiments. This region of about 200 bp (atr1−667 to −481) carries two copies of the predicted motif, named UAS-1 and UAS-2. S. cerevisiae cells that express the Rua1-AD fusion protein were transformed with reporter plasmids containing either the wild-type version of the 200-bp fragment derived from the atr1 promoter or mutated versions carrying mutations within the core element of the predicted motif, named UAS-1* and UAS-2* (Fig. 5B). The mutated UAS elements were tested either singly or jointly. To quantify the amount of transcription of the lacZ gene, we measured β-galactosidase activity in cultures of the transformed cells. The results are shown in Fig. 5C and revealed that strong β-galactosidase activity could be observed only when both wild-type motifs were present. Nucleotide exchanges within one of the predicted motifs resulted in strongly reduced β-galactosidase activity while no β-galactosidase activity was detected in cells transformed with the DNA fragment carrying mutations in both motifs. Taken together, these results confirm that the predicted sequence motif acts as a binding site for the C2H2 transcription factor Rua1, which acts as activator for transcription of the respective cluster genes.
DISCUSSION
We have identified the C2H2 zinc finger transcription factor Rua1, which is both necessary and sufficient to trigger expression of the biosynthetic gene cluster for the cellobiose lipid UA. The rua1 gene itself is part of the cluster and is located at its leftmost end. Since nitrogen limitation induces expression of the UA biosynthesis gene cluster, we assume that this regulation is mediated by Rua1, most probably by nitrogen-dependent expression of Rua1. We showed that Rua1 localizes to the nucleus and that it recognizes a conserved sequence element found in the promoter sequences of all cluster genes, with the only exception being the rua1 gene itself. While deletion of rua1 abolished transcription of all cluster genes tested, constitutive expression of rua1 resulted in constitutive UA production even under noninducing conditions, i.e., in the presence of a rich nitrogen source (Fig. 3A).
We used the yeast one-hybrid system to demonstrate that Rua1 is able to activate gene expression in the heterologous yeast system. This strongly suggests that regulation of transcription by Rua1 occurs by direct binding to the conserved sequence elements and does not require other transcription factors or additional U. maydis proteins. Rua1 is both necessary and sufficient to trigger UA biosynthesis. Thus, environmental and nutrient regulation of UA biosynthesis is presumably directly mediated by the level of Rua1 present in the cell. This would predict that all environmental signals that affect UA biosynthesis are integrated at the rua1 promoter. The rua1 gene is characterized by an unusually long promoter region spanning more than 4.7 kb. This region could accommodate binding sites for a number of different transcription factors involved in environmental and nutritional regulation of rua1 expression. However, one cannot exclude that rua1 expression might also be subject to posttranscriptional control, as was described for the global nitrogen regulator AreA in A. nidulans. The AreA transcript is specifically degraded in response to intracellular glutamine (6). One could imagine that this is also true for Rua1. Under inducing conditions, the rua1 transcript is stabilized, which results in UA biosynthesis. In contrast, the presence of a favored nitrogen source would result in transcript degradation. To distinguish between these potential regulatory schemes, the level of Rua1 expression should be followed by fusion of the promoter region to a reporter gene.
Truncation analysis of Rua1 indicated that the two zinc finger domains located at the C-terminal region are responsible for DNA binding. Interestingly, the amino acid sequences of both zinc finger domains deviate significantly from the C2H2 consensus sequence (34) and are not detected by database searches. This indicates that the zinc finger domain in Rua1, together with its relatives in other fungal species (Fig. 1), constitutes a novel fungus-specific subfamily of zinc finger transcription factors. Since some of these members have been identified in previous genetic screens but were annotated as proteins with unknown function, this finding sheds new light on their potential regulatory function. A prominent example is the meiotically upregulated gene meu26 in S. pombe (31), which could now be regarded as a potential regulator of meiosis-specific genes.
Our data strongly suggest that DNA binding to the conserved motif within the promoter regions of the cluster genes is mediated by the zinc finger domains. This is supported by the point mutation located within the zinc finger region in the rua1C747R mutant strain, which is unable to secrete UA anymore. This mutation affects a conserved cysteine residue located directly adjacent to one of the histidines putatively involved in the coordination of zinc. For the regulatory gene areA, mediating nitrogen metabolite repression in A. nidulans, it was shown that a single amino acid exchange within the zinc finger loop at position 102 (Leu to Val) also leads to derepression of genes involved in nitrogen metabolism (20). Interestingly, in the rua1C747R point mutant, expression of rua1 itself was affected, indicating that there might be some kind of autoregulation.
How expression of Rua1 is regulated remains to be elucidated. We suppose that regulatory proteins that sense global and specific nitrogen availability are involved. Candidate proteins for this regulation are the U. maydis AreA/NIT2 homolog Um10417 and the homolog of the pathway-specific activator NIT4, Um02808 (14). Alternatively, there might exist additional transcription factors that are specifically involved in the nutrient control of secondary metabolism genes. Initial experimental data indicate that nitrogen-dependent transcription of Rua1 is also regulated by another zinc finger transcription factor (Um02717), which we have termed Nit1 (B. Teichmann, L. Liu, and M. Bölker, unpublished data). It remains to be elucidated whether Nit1 is directly involved in nitrogen sensing. Interestingly, its deletion affects not only UA biosynthesis but also the production of MEL glycolipids.
The cis-acting element which acts as an upstream activating sequence and confers Rua1-specific transcription was identified by bioinformatic and mutational analysis. In the promoters of all structural genes involved in UA biosynthesis, at least one copy of the consensus sequence (T/G)(G/T)CGCAT(A/T)(C/T)(C/T)(G/A) was found. Interestingly, introduction of single point mutations in the tandem element upstream of the atr1 gene resulted in loss of activation, indicating that both UASs act synergistically in gene activation in vivo. A similar observation has already been described for binding sites of AflR, the transcriptional factor of the A. nidulans sterigmatocystin biosynthesis gene cluster. Deletion of only one of the three AflR binding sites within the stcU promoter results in decreased gene expression (7).
The secreted cellobiose lipid UA has broad antibacterial and antifungal spectra (11). Coinoculation of wild-type U. maydis cells effectively prevents infection of tomato leaves by Botrytis cinerea, the causal agent of gray mold disease. Therefore, it has been proposed that U. maydis could be used as a biocontrol agent. UA production plays an essential role for this antagonistic activity since mutants that are unable to produce UA do not prevent B. cinerea infections (29). Interestingly, a similar glycolipid is produced by the basidiomycetous fungus Pseudozyma flocculosa, which is a natural inhabitant of the phyllosphere. P. flocculosa has already been described as a potent biocontrol agent against fungal pathogens of plants (1, 10). The elucidation of the regulation of the UA biosynthesis gene cluster in U. maydis may therefore also open new avenues for studying and improving biocontrol activity in the highly related fungus P. flocculosa.
Secondary metabolite genes are often found to be clustered and are in general not constitutively expressed (18, 30). To understand the role of transcription factors in cluster regulation, those involved in secondary metabolism in Aspergillus strains were classified into different groups (15). AflR was the first member of a class of pathway-specific transcription factors. The criteria by which AflR was sorted into this class also apply to Rua1: Rua1 recognizes and binds to a specific sequence found in all promoters of the UA biosynthesis genes. There is evidence of self-regulation of Rua1 because a point mutation in the rua1 gene leads to a downregulation of itself, suggesting that Rua1 activates its transcription of its own gene. Deletion of rua1 abolishes expression of UA cluster genes, whereas constitutive expression of rua1 results in production of the secondary metabolite UA independently of nitrogen limitation. Thus, Rua1 acts as a central, highly pathway-specific transcription factor for UA biosynthesis. Secondary metabolites are often produced only under specific environmental conditions. The presence of a central regulator confers the advantage of allowing quick adaptation of cluster expression to different conditions by just changing the expression pattern of the pathway-specific transcription factor.
Acknowledgments
We thank Johannes Freitag for helpful discussions and Richard R. Bélanger for critical reading of the manuscript.
This work was supported by grants BO 2094/3-1 and GK1216 from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Avis, T. J., and R. R. Bélanger. 2002. Mechanisms and means of detection of biocontrol activity of Pseudozyma yeasts against plant-pathogenic fungi. FEMS Yeast Res. 2:5-8. [DOI] [PubMed] [Google Scholar]
- 2.Banuett, F., and I. Herskowitz. 1989. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. U. S. A. 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bok, J. W., D. Chung, S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, K. A. Kirby, and N. P. Keller. 2006. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 74:6761-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boothroyd, B., J. A. Thorn, and R. H. Haskins. 1956. Biochemistry of the Ustilaginales: XII. Characterization of extracellular glycolipids produced by Ustilago sp. Can. J. Biochem. Physiol. 34:10-14. [PubMed] [Google Scholar]
- 5.Brachmann, A., J. König, C. Julius, and M. Feldbrügge. 2004. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272:216-226. [DOI] [PubMed] [Google Scholar]
- 6.Caddick, M. X., M. G. Jones, J. M. van Tonder, H. Le Cordier, F. Narendja, J. Strauss, and I. Y. Morozov. 2006. Opposing signals differentially regulate transcript stability in Aspergillus nidulans. Mol. Microbiol. 62:509-519. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes, M., N. P. Keller, and T. H. Adams. 1998. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28:1355-1365. [DOI] [PubMed] [Google Scholar]
- 8.Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48:1035-1046. [DOI] [PubMed] [Google Scholar]
- 9.Fluharty, A. L., and J. S. O'Brien. 1969. A mannose- and erythritol-containing glycolipid from Ustilago maydis. Biochemistry 8:2627-2632. [DOI] [PubMed] [Google Scholar]
- 10.Hajlaoui, M. R., N. Benhamou, and R. R. Bélanger. 1992. Cytochemical Study of the antagonistic activity of Sporothrix flocculosa on rose powdery mildew, Sphaerotheca pannosa var. rosae. Phytopathology 82:583-589. [Google Scholar]
- 11.Haskins, R. H., and J. A. Thorn. 1951. Biochemistry of the ustilaginales: VII. Antibiotic activity of ustilagic acid. Can. J. Bot. 29:585-592. [Google Scholar]
- 12.Hewald, S., K. Josephs, and M. Bölker. 2005. Genetic analysis of biosurfactant production in Ustilago maydis. Appl. Environ. Microbiol. 71:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewald, S., U. Linne, M. Scherer, M. A. Marahiel, J. Kämper, and M. Bölker. 2006. Identification of a gene cluster for biosynthesis of mannosylerythritol lipids in the basidiomycetous fungus Ustilago maydis. Appl. Environ. Microbiol. 72:5469-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, E. C., M. J. Cahill, and B. J. Saville. 2007. Gene discovery and transcript analyses in the corn smut pathogen Ustilago maydis: expressed sequence tag and genome sequence comparison. BMC Genomics 8:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmeister, D., and N. P. Keller. 2007. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24:393-416. [DOI] [PubMed] [Google Scholar]
- 16.Jansen, G., C. Wu, B. Schade, D. Y. Thomas, and M. Whiteway. 2005. Drag&Drop cloning in yeast. Gene 344:43-51. [DOI] [PubMed] [Google Scholar]
- 17.Kämper, J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271:103-110. [DOI] [PubMed] [Google Scholar]
- 18.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 19.Kitamoto, D., H. Isoda, and T. Nakahara. 2002. Functions and potential applications of glycolipid biosurfactants—from energy-saving materials to gene delivery carriers. J. Biosci. Bioeng. 94:187-201. [DOI] [PubMed] [Google Scholar]
- 20.Kudla, B., M. X. Caddick, T. Langdon, N. M. Martinez-Rossi, C. F. Bennett, S. Sibley, R. W. Davies, and H. N. Arst, Jr. 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leveleki, L., M. Mahlert, B. Sandrock, and M. Bölker. 2004. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 54:396-406. [DOI] [PubMed] [Google Scholar]
- 22.Massiah, M. A., J. A. Matts, K. M. Short, B. N. Simmons, S. Singireddy, Z. Yi, and T. C. Cox. 2007. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J. Mol. Biol. 369:1-10. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schäfer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 27.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 28.Spellig, T., A. Bottin, and R. Kahmann. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503-509. [DOI] [PubMed] [Google Scholar]
- 29.Teichmann, B., U. Linne, S. Hewald, M. A. Marahiel, and M. Bölker. 2007. A biosynthetic gene cluster for a secreted cellobiose lipid with antifungal activity from Ustilago maydis. Mol. Microbiol. 66:525-533. [DOI] [PubMed] [Google Scholar]
- 30.Walton, J. D. 2000. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 30:167-171. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, T., K. Miyashita, T. T. Saito, T. Yoneki, Y. Kakihara, K. Nabeshima, Y. A. Kishi, C. Shimoda, and H. Nojima. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29:2327-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, T. E., T. J. Fahrner, M. Johnston, and J. Milbrandt. 1991. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252:1296-1300. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, T. E., K. A. Padgett, M. Johnston, and J. Milbrandt. 1993. A genetic method for defining DNA-binding domains: application to the nuclear receptor NGFI-B. Proc. Natl. Acad. Sci. U. S. A. 90:9186-9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfe, S. A., L. Nekludova, and C. O. Pabo. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29:183-212. [DOI] [PubMed] [Google Scholar]
- 35.Woloshuk, C. P., K. R. Foutz, J. F. Brewer, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]