Abstract
Cronobacter spp. are opportunistic food-borne pathogens that can cause severe and sometimes lethal infections in neonates. In some outbreaks, the sources of infection were traced to contaminated powdered infant formula (PIF) or contaminated utensils used for PIF reconstitution. In this study, we investigated biofilm formation in Cronobacter sakazakii strain ES5. To investigate the genetic basis of biofilm formation in Cronobacter on abiotic surfaces, we screened a library of random transposon mutants of strain ES5 for reduced biofilm formation using a polystyrene microtiter assay. Genetic characterization of the mutants led to identification of genes that are associated with cellulose biosynthesis and flagellar structure and biosynthesis and genes involved in basic cellular processes and virulence, as well as several genes whose functions are currently unknown. In two of the mutants, hypothetical proteins ESA_00281 and ESA_00282 had a strong impact on flow cell biofilm architecture, and their contribution to biofilm formation was confirmed by genetic complementation. In addition, adhesion of selected biofilm formation mutants to Caco-2 intestinal epithelial cells was investigated. Our findings suggest that flagella and hypothetical proteins ESA_00281 and ESA_00282, but not cellulose, contribute to adhesion of Cronobacter to this biotic surface.
Biofilms are interface-associated consortia of microorganisms that are typically embedded in an endogenous slimy matrix referred to as extracellular polymeric substance (EPS). It is generally accepted that growth as a biofilm is the predominant microbial lifestyle in nature. Biofilms have several phenotypic characteristics that clearly set them apart from planktonic cultures, most notably increased resistance to a variety of environmental influences (16), which makes their eradication more difficult. Microbial biofilms are of special concern to the food industry, as biofilms on raw materials or food contact surfaces represent possible sources of product contamination with spoilage or pathogenic microorganisms (for a recent review, see reference 4).
Cronobacter spp. are opportunistic food-borne pathogens that can cause severe disease in neonates which may present as septicemia, meningitis, or necrotizing enterocolitis (NEC). In several outbreaks, the source of infection was traced to contaminated powdered infant formula (PIF) or to spoons and blenders used in preparation of PIF (8, 10). The genus Cronobacter currently comprises six species: Cronobacter sakazakii, Cronobacter dublinensis, Cronobacter turicensis, Cronobacter malonaticus, Cronobacter muytjensii, and Cronobacter genomospecies 1 (20). Cronobacter spp. display remarkable resistance to desiccation compared to other Enterobacteriaceae (7), which may contribute to their long-term survival in PIF and on surfaces. Few studies of biofilm formation by Cronobacter spp. have been conducted so far. It has been observed that some strains are able to form biofilms on glass, stainless steel, polyvinyl chloride (PVC), polycarbonate, silicone, and enteral feeding tubes in different media (19, 24, 28). Like biofilm formation in other bacteria, biofilm formation is different for different strains and is highly dependent on the medium and surface used. Furthermore, the survival of C. sakazakii in biofilms under different environmental conditions has been investigated (23), and increased resistance of Cronobacter biofilms to disinfectants has been demonstrated (25). Cellulose has been described as a component of the Cronobacter extracellular matrix (15, 28, 51).
In this study, we performed a genetic analysis of biofilm formation by Cronobacter sakazakii strain ES5, a clinical isolate, by using random transposon mutagenesis and subsequent screening of a mutant library for altered biofilm phenotype using a microtiter assay system. In addition, the biofilm structure of the wild type and selected mutants in a continuous-culture flow cell system was investigated by using confocal laser scanning microscopy (CLSM). Finally, we tested whether for selected mutants the defects in biofilm formation observed on the abiotic surface had an influence on the capacity of C. sakazakii to adhere to Caco-2 intestinal epithelial cells.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids used in this study are listed in Tables 1 and 2. C. sakazakii ES5 is a clinical strain that exhibited the strongest biofilm formation on polystyrene surfaces in preliminary experiments performed with a panel of Cronobacter strains.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Reference or source |
|---|---|---|
| Cronobacter sakazakii ES5 | Human isolate, wild type | 28 |
| Escherichia coli strains | ||
| INVαF′ | F′ endA1 recA1 hsdR17(rk− mk+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169 λ | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids and transposons | ||
| pCR2.1 | Kmr Apr; TA cloning vector, lacZα | Invitrogen |
| pP1b | pUC18NotI derivative containing ESA_00281 and ESA00_280 | This study |
| pR8b | pUC18NotI derivative containing ESA_00282 | This study |
| pUC18NotI | Apr; cloning vector | 17 |
| EZ-Tn5 <KAN-2> (Tn) | Kmr; mini-Tn5 transposon | Epicentre |
TABLE 2.
Description of mutations in C. sakazakii ES5 that result in reduced biofilm on polystyrene
| Functional group | Mutant(s)a | Locus tag (gene ID no.)b | Annotationc |
Reference(s) | Phenotype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Description (organism) | Accession no. | No. of positive amino acids/total no. (%) | Biofilm in CV assay (% of wild type, mean ± SD)g | CR agar | Flagella (EM) | ||||
| Cellulose biosynthesis | BF3, BF10 | ESA_04207 (5550384) | bcsC (yhjL) | Cellulose synthase subunit BcsC (Cronobacter sakazakii BAA-894)b | YP_001440 223.1 | 1,167/1,167 (100) | 15, 38, 52 | 33.6 ± 25.4 | sawh | NDk |
| BF6 | ESA_04199 (5550446) | bcsG (yhjU) | Conserved hypothetical protein (Cronobacter sakazakii)d | CAM32322.1 | 557/561 (99) | 15, 38, 52 | 20.5 ± 8.9 | saw | ND | |
| BF12 | ESA_04204 (5550385) | bcsA (yhjO) | Cellulose synthase catalytic subunit (Cronobacter sakazakii BAA-894)b | YP_001440 220.1 | 872/872 (100) | 15, 38, 52 | 15.5 ± 5.9 | saw | ND | |
| Flagellar structure or biosynthesis | BF2 | ESA_01356 (5549172) | flhE | Flagellar protein FlhE (Cronobacter turicensis) | CBA31743.1 | 118/119 (99) | 43 | 19.7 ± 6.3 | pdari | Like wild type |
| BF5 | ESA_01287 (5549196) | fliD | Flagellar hook-associated 2 domain protein 2 (Cronobacter turicensis) | CBA31883.1 | 471/476 (98) | 29, 48 | 9.3 ± 5.1 | pdar | Shorter, brittle | |
| BF13 | ESA_02266 (5551730) | flgJ | Peptidoglycan hydrolase (Cronobacter sakazakii BAA-894)b | YP_001438 351.1 | 321/321 (100) | 29 | 77.9 ± 6.4 | pdar | Absent | |
| Basic cellular processes | ND | |||||||||
| Cell division | BF1 | ESA_02449 (5549447) | ftsK | DNA translocase FtsK (Cronobacter turicensis) | CBA29614.1 | 1,264/1,364 (92) | 6 | 8.3 ± 2.6 | pdar | ND |
| Energy production | BF8 | ESA_02873 (5548784) | cyoD | Cytochrome o ubiquinol oxidase protein CyoD (Cronobacter turicensis) | CBA28615.1 | 109/109 (100) | 39 | 19.3 ± 6.1 | pdar | ND |
| BF9 | ESA_00861 (5550827) | alsS (ilvB) | Acetolactate synthase (Cronobacter sakazakii BAA-894)b | YP_001436 968.1 | 559/559 (100) | 36 | 16.5 ± 3.4 | pdar | ND | |
| Possibly involved in virulence | BF17 | ESA_pESA3p05536 (5552640) | mgtB | Magnesium-transporting ATPase, P-type 1 (Cronobacter turicensis) | CBA34629.1 | 853/904 (94) | 41 | 22.5 ± 9.1 | pdar | ND |
| Unknown function | BF4 | ESA_04103 (5551795) | NAe | Hypothetical protein Ent638_0111 (Enterobacter sp. 638) | ABP58801.1 | 361/425 (84) | NA | 66.9 ± 20.1 | pdar | Like wild type |
| BF11, BF15 | ESA_00282 (5549918) | NA | Hypothetical protein (Cronobacter turicensis) | CBA33797.1 | 164/166 (98) | NA | 72.8 ± 21.3 | pdarsej | Like wild type | |
| BF14 | ESA_00281 (5549972) | NA | Hypothetical protein (Cronobacter turicensis) | CBA33799.1.1 | 132/133 (99) | NA | 60.5 ± 36.9 | pdarse | Like wild type | |
| BF16 | NA | NA | Similar to Escherichia coli plasmids pMG828-5, p6148, and pE2348f | DQ995355.1, EU580136.1, and FM180570.1 | NA | NA | 34.3 ± 24.0 | pdar | ND | |
Transposon-containing derivative(s) of C. sakazakii ES5.
Based on the genome of C. sakazakii BAA-894 (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=12720).
Based on a BlastP search with BAA-894 protein sequences of the NCBI nonredundant protein sequences on 15 January 2010.
C. sakazakii ES5 sequence from BAC clone (15).
NA, not applicable.
Based on BlastN search with the sequence obtained for ES5 mutant of the nucleotide collection (nr/nt) on 15 January 2010.
The value for the wild-type biofilm in the CV assay was 100% ± 9.2%.
saw, smooth and white (38).
pdar, pink, dry, and rough (38).
pdarse, pink, dry, and rough with smooth edge (this study).
ND, not determined.
Media and growth conditions.
Medium ingredients were obtained from BD (Franklin Lakes, NJ), Fluka (Buchs, Switzerland), Merck (Darmstadt, Germany), and Sigma (Buchs, Switzerland). C. sakazakii and Escherichia coli were routinely grown in modified LB broth (3) at 37°C with vigorous agitation, unless otherwise stated. Media were solidified by addition of 12 g/liter agar. When required, antibiotics were added to the following final concentrations: ampicillin, 100 μg/ml; and kanamycin, 50 μg/ml. For blue-white selection, 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-1-thiogalactopyranoside were added to final concentrations of 40 μg/ml and 0.2 mM, respectively. Congo red (CR) agar and calcofluor (Fluorescent brightener 28) agar were prepared as described by Römling et al. (38) and Grimm et al. (15), respectively.
Growth in liquid cultures was monitored by measuring the optical density at 600 nm (OD600) with an Ultrospec II spectrophotometer (Biochrom, Cambridge, United Kingdom) or by counting viable cells in 10-fold serial dilutions on plate count agar (Oxoid, Basingstoke, United Kingdom).
CV microtiter biofilm assay.
Quantification of biofilms grown in microtiter dishes was performed as described by Huber et al. (18). AB minimal medium [2 g/liter (NH4)2SO4, 6 g/liter Na2HPO4, 3 g/liter KH2PO4, 3 g/liter NaCl, 2 mM MgCl2, 0.1 mM CaCl2, 3 μM FeCl3·6H2O) supplemented with 0.4% (wt/vol) maltose was used as the growth medium, and cultures were incubated at 37°C for 45 h. After crystal violet (CV) staining, the absorbance at 570 nm was measured using a BioTek Synergy HT microplate reader (BioTek Instruments, Inc., Winooski, VT).
Construction of the transposon mutant library.
A library of random transposon mutants of C. sakazakii strain ES5 was constructed using an EZ-Tn5 <KAN-2>Tnp Transposome kit (Epicentre, Madison, WI) by following the manufacturer's instructions. Mutants were picked to wells of polystyrene microtiter dishes (Nunc, Denmark) containing modified LB medium supplemented with 7.5% glycerol and 50 μg/ml kanamycin, grown overnight at 37°C, and stored at −20°C.
DNA extraction, cloning, transformation, and sequencing.
All kits used for DNA purification were obtained from Qiagen (Hilden, Germany) and were handled according to the manufacturer's instructions. Unless otherwise stated, chromosomal DNA was purified using a DNeasy blood and tissue kit. Plasmids were extracted with QIAprep spin miniprep or plasmid midi kits. DNA fragments obtained from PCRs, restriction digests, and agarose gels were purified using MinElute PCR cleanup and MinElute gel purification kits. Concentrations of nucleic acids were determined using a Nanodrop ND-1000 UV/visible spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Cloning, restriction analysis, and transformation of C. sakazakii and E. coli were performed essentially as described by Sambrook et al. (40). Restriction enzymes and corresponding buffers were obtained from Roche (Mannheim, Germany), and T4 DNA ligase was obtained from New England Biolabs (Ipswich, MA). All sequencing was performed by Microsynth (Balgach, Switzerland).
Southern analysis and identification of transposon insertion sites.
Southern analysis was performed as described previously (21). For identification of insertion sites by the subcloning procedure, chromosomal DNA of the transposon mutants was digested with SphI, and the fragments were ligated into pUC18NotI (Table 1) digested with the same enzyme. Each ligation mixture was electroporated into E. coli XL1-Blue, and transformants carrying a plasmid containing the transposon sequence were selected on modified LB medium containing 50 μg/ml kanamycin. Plasmids were extracted from the selected clones, and transposon-flanking regions were sequenced with primer KAN-2 FP1 (Table 3).
TABLE 3.
Primers used in this study
| Primer | Nucleotide sequence (5′→3′)a | Reference or source |
|---|---|---|
| 280r4 | CGG GCC AAG CTT GAA CGC TGG ATC AGG TGA | This study |
| 281f1 | CCG GGC AAG CTT TAC TGT TCA TGC GCA GGC | This study |
| 282f2 | CCG GGC AAG CTT GGA ATA CGA TAA CGC GCC | This study |
| 282r1 | CGG GGC AAG CTT AAG CGT TCC ATA GCG TCC | This study |
| KAN-2 FP1 | ACC TAC AAC AAA GCT CTC ATC AAC C | Epicentre |
| M13 | TGT AAA ACG ACG GCC AG | New England Biolabs |
| M13r | CAG GAA ACA GCT ATG ACC | New England Biolabs |
| Tn5PCRF | GCT GAG TTG AAG GAT CAG ATC | 44 |
Recognition sequences of restriction enzymes are underlined.
For single-primer PCR, DNA fragments were amplified from chromosomal DNA of the transposon mutants using primer Tn5PCRF (Table 3) with the GoTaq Green PCR system (Promega, Madison, WI) and the reaction conditions described by Karlyshev et al. (22). PCR products were purified and used as templates in a sequencing reaction using primer KAN-2 FP2.
Transposon insertion sites were determined by performing a BlastN search (1) with the genome of C. sakazakii BAA-894 (accession numbers NC_009778, NC_009779, and NC_009780) or the NCBI nucleotide collection (nr/nt). Missing annotations or gene names were added manually by performing a BlastX (1) search with the sequence obtained from the transposon mutants, backed up by a BlastP (1, 2) search with the corresponding open reading frame (ORF) sequence of BAA-894 with the NCBI nonredundant protein sequences. All Blast searches were carried out using default parameters. The cellular localizations of the affected proteins were predicted by Psortb v.2.0 (12) and cello (50).
Construction of complementation plasmids.
The putative ESA_00281-ESA_00280 operon and the ESA_00282 ORF, both including flanking sequences, were amplified from wild-type chromosomal DNA using primer pairs 281f1/280r4 and 282f2/282r1, respectively. PCR products were ligated into pCR2.1 (Table 1) using a TA cloning kit (Invitrogen, Carlsbad, CA), and inserts were verified by sequencing using primers M13 and M13r (Table 3). Inserts were recovered from the plasmids by HindIII or BamHI (Roche, Mannheim, Germany) digestion, gel purified, and cloned into pUC18NotI digested with the same enzyme to obtain plasmids pP1b and pR8b.
Electron microscopy.
Bacteria were grown in liquid cultures overnight at 37°C with agitation in AB minimal medium supplemented with 0.8% lactose and were examined by electron microscopy (EM) as previously described (47).
Cultivation of biofilms in flow cells and confocal laser scanning microscopy (CLSM).
Biofilms were grown in flow cells containing 24- by 50-mm no. 1 glass coverslips (Menzel Gläser, Braunschweig, Germany) as the substratum in modified LB medium diluted 1:20 with distilled water. Ampicillin (100 μg/ml) was added to the medium when it was required. Flow cell system components were obtained from DTU (Lyngby, Denmark) (flow cells and bubble traps) and Omnilab (Mettmenstetten, Switzerland) (silicone tubes). The flow cell system was operated with a Watson-Marlow 205S peristaltic pump (Watson-Marlow, Wilmington, MA) at a flow rate of 0.7 mm/s.
Bacteria were subcultured from fresh overnight cultures in modified LB medium for 4 h at 37°C with shaking before they were diluted in 0.85% NaCl to obtain an OD600 of 0.1. Flow cell channels were inoculated in duplicate with 350 to 400 μl of a cell suspension, and cells were allowed to attach for 1 h at room temperature without a flow. After this, the flow cell system was incubated at 30°C with a constant flow. At 24 h or 45 h after start of incubation, flow cells were rinsed with 0.85% NaCl for 20 min. To stain the biofilms for CLSM, 350 to 400 μl of a 10 nM SYTO 9 solution (Invitrogen, Carlsbad, CA) was injected into the flow cell channels and incubated for 15 min in the dark. After 10 min of rinsing with 0.85% NaCl, biofilms were observed with a Leica TCS SPE confocal microscope (Leica Microsystems, Wetzlar, Germany) at a magnification of ×400. Image scans were performed using a 488-nm argon laser for excitation and a 490- to 690-nm emission band for detection. At least two representative image stacks were captured for each flow cell channel.
Adhesion to Caco-2 cells, Giemsa staining, and microscopy of cell monolayers.
Caco-2 intestinal epithelial cells were routinely maintained in Gibco minimal essential medium (MEM) containing 100 μM MEM nonessential amino acids, 4 mM GlutaMAX-I, 10% (vol/vol) fetal calf serum (FCS), and 200 μg/ml gentamicin at 37°C in a 5% CO2 atmosphere. All cell culture media, additives, and phosphate-buffered saline (PBS) were obtained from Invitrogen (Carlsbad, CA), except for fetal calf serum, which was obtained from PAA (Pasching, Austria). To assess the adhesion of C. sakazakii ES5 and derivatives of this strain to intestinal epithelial cells, 105 Caco-2 cells/well were seeded in 24-well cell culture dishes (TPP, Trasadingen, Switzerland) containing 1 ml test medium (Gibco Iscove's modified Dulbecco's medium supplemented with 10% [vol/vol] FCS) per well and allowed to grow for 72 h at 37°C in the presence of 5% CO2. Bacteria from fresh overnight cultures were subcultured in modified LB medium for 4 h at 37°C with agitation and subsequently diluted in test medium to obtain a concentration of 107 CFU/ml. Caco-2 cells were infected by replacing the medium with 1 ml of a bacterial suspension, which resulted in an infection dose of 107 bacteria/well (multiplicity of infection [MOI], approximately 10:1). Infected cells were incubated for 3 h at 37°C in the presence of 5% CO2. After five washes with 1 ml PBS/well, Caco-2 cells were lysed by addition of 1 ml 0.2% Triton X-100 in PBS/well and subsequent incubation at 37°C for 30 min. Total cell-associated bacteria were enumerated by serially diluting the lysates in PBS and plating them on plate count agar.
Giemsa staining of Caco-2 monolayers was performed as previously described (30), and slides were examined at a magnification of ×1,000 using an Olympus VANOX-S microscope (Olympus, Hamburg, Germany) fitted with a Zeiss Axiocam digital camera using the Zeiss Axioview software (Carl Zeiss AG, Germany).
Statistic analysis.
All comparisons were performed using a ranked t test for unequal variances (Welch test) using a script for the statistical computing software R (35).
RESULTS
Isolation of mutants with altered biofilm formation.
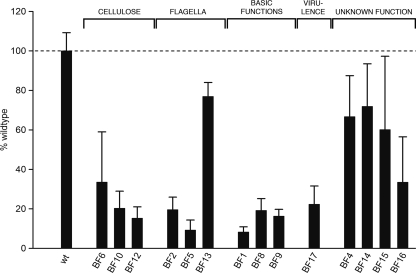
A C. sakazakii strain ES5 random transposon mutant library comprising 3,328 clones was generated. These clones were screened for a reduced-biofilm phenotype on polystyrene using a crystal violet microtiter assay. Only mutants that exhibited the wild-type growth phenotype but a >15% reduction in biofilm formation were characterized genetically. Based on these criteria, 21 transposon mutants with impaired biofilm formation (Fig. 1 and Table 2) were selected.
FIG. 1.
Quantification of biofilm. Biofilms of wild-type strain ES5 and mutants were measured using the CV microtiter assay. The values are means from at least two independent experiments with 2 to 8 replicates in each experiment. The error bars indicate one standard deviation. The dashed line indicates the wild-type strain ES5 level (100%). All mutant values were significantly different from the wild-type value (P < 0.001, ranked Welch test). wt, wild type.
Genetic characterization of the isolated mutants.
Southern hybridization confirmed that only one transposon was present in all of the mutants.
Transposon insertion sites were determined using two different approaches: a subcloning procedure and single-primer PCR if the subcloning procedure failed to determine the sites. Using standard homology searches, the transposon insertion sites were mapped in the genome of strain C. sakazakii BAA-894, and functions were assigned to the defective genes. While 16 of the mutations identified affected genes located on the chromosome of strain BAA-894, 3 mutations were found to be in sequences that are plasmid borne in BAA-894 or other organisms. The genes with transposon insertions were grouped into functional classes, as shown in Table 2. Mutations were found in genes involved in cellulose biosynthesis, motility, basic cellular functions, and virulence and in genes whose functions are unknown. The following two groups of “sibling” mutants with identical transposon insertion sites were discovered: three mutants carrying the transposon in the flhE gene (identical to mutant BF2) and two mutants carrying the transposon in a putatively plasmid-encoded sequence (representative mutant BF16). In two pairs of mutants (mutants BF3 and BF10 and mutants BF11 and BF15), the same gene was mutated, but at different positions.
In total, four mutants were defective in the cellulose biosynthesis pathway. The biofilm formation by these mutants was 15 to 34% of the biofilm formation by the wild type, as quantified by the CV microtiter assay (Fig. 1 and Table 2).
One mutant (BF12) carried the transposon in bcsA, which codes for the catalytic subunit of the cellulose synthase. Two nonisogenic mutants were defective in bcsC (BF3 and BF10), which encodes the cellulose biosynthesis operon protein C, a putative oxidoreductase. Another mutant was defective in bcsG, whose product has not been characterized yet.
Five mutants were defective in flagellum-associated genes. In one mutant (BF5), the transposon was inserted into fliD, which encodes the flagellar capping protein, and in another mutant (BF13) the transposon was inserted into flgJ, a muramidase gene involved in the biosynthesis of the flagellum. In the three remaining mutants (identical to BF2), the transposon was found to be in a flagellar gene (flhE) whose function is still unknown, at the same insertion position. For the fliD and flhE mutants, the amount of biofilm in the CV assay was dramatically reduced (9% and 20% of the wild-type amount), while for the flgJ mutant the amount of biofilm was 84% of the wild-type amount.
In three mutants, the transposon was inserted into genes that are involved in fundamental cellular processes, including cell division (ftsK, involved in chromosome segregation [mutant BF1]) and energy metabolism (cyoD, fourth subunit of the cytochrome o ubiquinol oxidase [mutant BF8], and alsS, acetolactate synthase gene involved in mixed acid fermentation [mutant BF9]). As these mutations cause severe cellular defects or retarded growth due to impaired energy metabolism, the observed reductions in biofilm formation compared to the wild type (93%, 81%, and 82% reductions, respectively) have to be considered nonspecific in the context of biofilm formation.
In mutant BF17, the transposon was inserted into mgtB, a gene encoding a P-type Mg2+ transport ATPase that has been described in the context of invasion of epithelial cells and macrophages. This gene is in a plasmid in C. sakazakii BAA-894. Disruption of mgtB resulted in 77% less biofilm than the amount of biofilm obtained with the wild type in the CV microtiter assay.
Six mutants were affected in hypothetical proteins. One mutant (BF4) carried the transposon in ORF ESA_04103, whose product is a hypothetical protein with an unknown function. The biofilm produced by this mutant had a markedly different appearance when it was assessed visually in the microtiter assay. This translated into a 31% reduction in CV quantification compared to the wild type. Only one similar sequence, a sequence from Enterobacter sp. 638, was found by a BlastP search using the NCBI nonredundant protein database. The subcellular localization of this protein was predicted to be in the inner membrane. The biofilm architecture of mutant BF4 was investigated further using the flow cell system. However, no difference from the wild type was observed (data not shown).
Two identical mutants (represented by BF16) were isolated in which the transposon insertion site could not be mapped to any sequence in the genome of BAA-894. However, the nucleotide sequence obtained for the mutants was 89 to 90% similar to the nucleotide sequences of certain cryptic plasmids of different E. coli strains (Table 2). In two of these plasmids (pMG828-5 and p6148), the insertion site was located in intergenic regions, while in plasmid pE2348-2 the insertion site was mapped to a putative ORF encoding a hypothetical protein. The defect in this putatively plasmid-borne region led to a 66% reduction in the amount of biofilm compared to the amount of biofilm produced by the wild type in the CV microtiter assay (Fig. 1 and Table 2).
Finally, three mutants were found to carry the transposon in two other genes encoding hypothetical proteins, ORFs ESA_00281 and ESA_00282. In one mutant (BF14) the transposon was inserted into ESA_00281, and in two mutants (BF11 and BF 15) it was inserted into ESA_00282, but at different sites. The mutant biofilms had an appearance in the microtiter assay that was clearly different and bound 73% (mutant BF14) or 60% (mutant BF15) of the amount of CV trapped by the wild-type biofilm (Fig. 1 and Table 2). The mutated genes are located next to each other in the genome of C. sakazakii BAA-894, but they are oriented divergently. ORF ESA_00281 is organized in a putative operon with ORF ESA_00280, which has also not been characterized to date. Similar proteins are also encoded in the Salmonella, Enterobacter, and Citrobacter genomes sequenced so far. In E. coli strains the genes seem to be absent, except for O157:H7 strain EC4024.
Phenotypic characterization of the mutants.
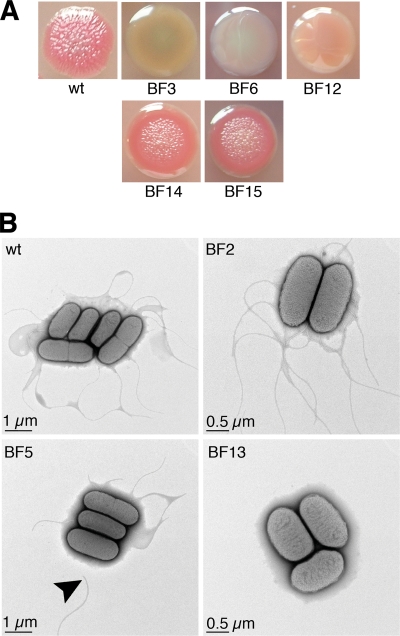
The mutants were characterized further using suitable phenotypic assays. As the extracellular matrix is known to play an important role in bacterial biofilm formation, the wild type and all mutants were streaked on CR agar and on calcofluor agar, both of which contain cellulose-binding dyes. The former medium also contains Coomassie brilliant blue to indicate proteinaceous components of the extracellular matrix. The wild type formed pink, rugose colonies similar to the pdar (pink, dry, and rough) morphotype defined for Salmonella, which indicates that cellulose is present but curli fimbriae are not present in the cellular matrix (38) (Fig. 2A). All mutants displayed the wild-type phenotype on both CR agar and calcofluor agar, except for mutants BF14 (ESA_00281::Tn), BF15 (ESA_00282::Tn), and, as expected, the four cellulose mutants (Table 2 and Fig. 2A). In mutants BF14 and BF15, an interesting phenotype was observed, which was the same for both mutants. The colonies were the same pink color as the wild-type colonies, but they had a smooth colony edge. The typical wrinkled structure observed in wild-type colonies on CR agar was restricted to the colony center (Fig. 2A). As expected, the cellulose mutants formed smooth, whitish colonies, a morphotype resembling the saw (smooth and white) morphotype of Salmonella colonies (38), which indicates that neither cellulose nor curli fimbriae were present (Fig. 2A). On calcofluor agar, the wild-type colonies were brightly fluorescent under UV light, indicating that cellulose was present. All of the mutants were equally fluorescent, except for the cellulose mutants, whose colonies showed only weak fluorescence (data not shown).
FIG. 2.
Confirmation of mutant phenotype. (A) Colony morphology of wild-type strain ES5, cellulose biosynthesis mutants BF3 (bcsC::Tn), BF6 (bcsF::Tn), and BF12(bcsA::Tn), and mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn) on CR agar. (B) Electron micrographs of wild-type strain ES5 and flagellum mutants BF2 (flhE::Tn), BF5 (fliD::Tn), and BF13 (flgJ::Tn). The arrowhead indicates an unattached flagellar fragment. wt, wild type.
Wild-type strain ES5, flagellar mutants BF2 (flhE::Tn), BF5 (fliD::Tn), and BF13 (flgJ::Tn), and mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn) were examined by electron microscopy to investigate flagellar structure. Examination of the flagellar motility of the wild type and the flagellar mutants was not successful due to the immobility of the wild-type strain under standard motility assay conditions, suggesting that swimming motility might not be critically important for formation of a C. sakazakii biofilm. EM analysis showed that wild-type strain ES5 was peritrichous, which is typical of most Enterobacteriaceae. However, the flgJ mutant (BF13) was aflagellate, and the fliD mutant (BF5) had shorter flagella that seemed to be more brittle, as unattached flagellum fragments were observed in the sample (Fig. 2B). No difference from the wild type was observed for mutants BF2, BF14, and BF15 with respect to structure or average number of flagella per cell.
Complementation of mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn).
Because of the interesting phenotypes and the adjacent localization of the mutant genes on the BAA-894 genome, we decided to perform genetic complementation experiments with the BF14 and BF15 mutants. Complemented mutants BF14(pP1b) and BF15(pR8b) and wild-type strain ES5(pUC18NotI) were first investigated using the CV microtiter assay. However, the presence of the high-copy-number cloning vector pUC18NotI alone reduced biofilm formation by wild-type strain ES5 by 43% (Fig. 3). The medium-copy-number plasmid pBBR1MCS (27) was also investigated as a possible alternative, but its negative effect on the wild-type biofilm was even greater, and there was a 60% reduction in biofilm formation compared to the wild type (data not shown). Therefore, we decided to use pUC18NotI for our complementation experiments. OD600 measurements indicated that the growth rates of the transformants were lower than that of the wild type (data not shown), possibly as a result of the increased metabolic load caused by the presence of a plasmid. Replication of the plasmid and constitutive expression of plasmid-borne antibiotic selection markers drain the cell's resources in terms of energy and building blocks (14).
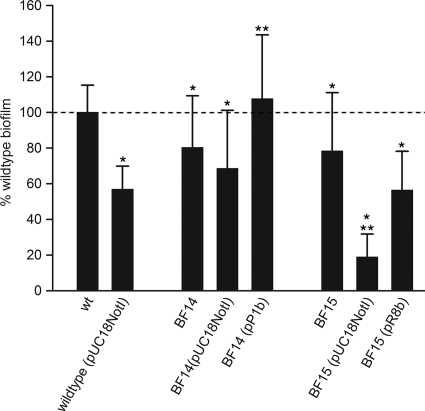
FIG. 3.
Complementation of mutants BF14 and BF15. Quantification of biofilm for wild-type strain ES5, strain ES5(pUC18NotI), mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn), and complemented mutants in the CV microtiter assay. The values are means of values from at least three independent experiments with at least six replicates in each experiment. The error bars indicate one standard deviation. The dashed line indicates the ES5(pUC18NotI) level. All values were compared to the wild-type level, and one asterisk indicates that there is a significant difference (P < 0.05, Welch test). BF14 (pUC18NotI), BF14(pP1b), BF15(pUC18NotI), and BF15(pR8b) were also compared to ES5(pUC18NotI). Significant differences (P < 0.01, Welch test) are indicated by two asterisks. wt, wild type.
Complemented mutants BF14(pP1b) and BF15(pR8b) were investigated by using CR agar, the CV microtiter assay, and the flow cell system. As controls, strain BF14(pUC18NotI), strain BF15(pUC18NotI), and wild-type strain ES5(pUC18NotI) were included in these experiments. In the CV microtiter assay, the presence of pUC18NotI reduced the amount of biofilm produced by mutant BF15 to 19% of the amount produced by the wild type, which corresponded to a further reduction of 60%. In mutant BF14, the plasmid-induced additional reduction was less pronounced; mutant BF14 produced 68% of the biofilm produced by the wild type in the presence of pUC18NotI, corresponding to a further reduction of 14%. Despite these effects, expression of ORFs ESA_00280 and ESA_00281 or ORF ESA_00282 from the complementation plasmids restored the levels of biofilm production, as quantified by the CV microtiter assay, to 108% and 56% of wild-type level, respectively, showing that in trans expression of the mutated genes clearly had a positive effect (Fig. 3). In the flow cell system (see below) and on CR agar, no effects of pUC18NotI were apparent in wild-type strain ES5 or mutants BF14 and BF15. The wild-type phenotype could be restored in both mutants by the presence of the corresponding complementation plasmids both on CR agar (data not shown) and in the flow cell system (Fig. 4B).
FIG. 4.
CLSM image stack profiles of flow cell biofilms. Each large panel shows a horizontal cross section of a flow cell-grown biofilm, while the smaller panels show vertical cross sections. The dashed lines in the panels indicate the planes of the other two cross sections. (A) C. sakazakii strain wild-type strain ES5 at 24 h and 45 h after start of incubation. BL, basal layer; MC, microcolony. (B) Mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn) and complemented mutants BF14 and BF15 at 45 h after the start of incubation. All images were taken at a magnification of ×400.
Biofilm architecture in a flow cell system.
Flagellar mutant BF2 (flhE::Tn) and three mutants in which uncharacterized genes were disrupted, BF4 (ESA_04103::Tn), BF14 (ESA_00281::Tn), and BF15 (ESA_00282::Tn), were characterized further by comparison of their biofilms with wild-type biofilms grown in a continuous-culture flow cell system. After 24 h of incubation, a basal layer of cells about 20 μm thick was observed for the wild type, and microcolonies protruded about 30 μm. After 45 h, the thickness of the basal layer of the wild-type biofilm was unchanged, but the height of microcolonies had increased slightly to about 40 μm (Fig. 4A).
For mutants BF2 (flhE::Tn) and BF4 (ESA_04103::Tn), no difference from the wild-type biofilm architecture was apparent (data not shown). However, the biofilm architecture of mutants BF14 (ESA_00281::Tn) and BF15 (ESA:00282::Tn) was different, and it was the same for both strains. After 24 h of incubation, only microcolonies were observed, which were not interconnected by a homogeneous layer of bacteria like the layer observed for the wild type at this stage. Only single attached cells were detected on the glass surface between the microcolonies (data not shown). After 45 h, clonal growth of the singly adherent bacteria and the bacteria in microcolonies seemed to have occurred; however, patches of uncolonized surface were still visible between the microcolonies (Fig. 4B), which was also in contrast to the findings for the wild type.
Adhesion of selected mutants to Caco-2 intestinal epithelial cells.
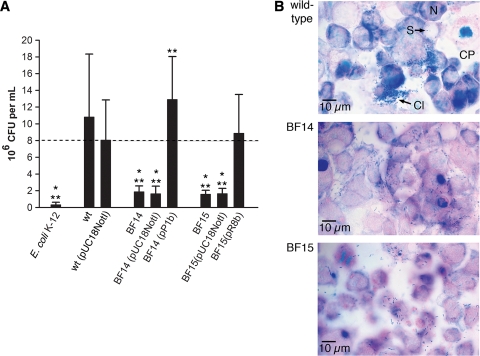
To investigate whether genes identified in the screening for defects in biofilm formation on polystyrene also play a role in adhesion to a biotic surface, we assessed the capacity of selected mutants to adhere to confluent monolayers of Caco-2 cells by measuring the total cell-associated bacteria after 3 h of incubation. The results are summarized in Fig. 5. The flhE mutant (BF2), the mgtB mutant (BF17), and the cellulose-negative bcsA mutant (BF12) were not different from the wild type (data not shown). In the aflagellate flgJ mutant (BF13) (data not shown), adhesion was reduced almost to the level of the negative control E. coli K-12, for which the number of bacteria recovered was approximately 2 orders of magnitude lower than the number of wild-type bacteria recovered. For the ESA_00281 and ESA_00282 mutants BF14 and BF15 the numbers of bacteria recovered from the Caco-2 cells were 1 order of magnitude less than the numbers of the wild-type bacteria recovered. The total numbers of cell-associated bacteria were also determined for mutants BF14 and BF15 harboring either pUC18NotI or complementation plasmids pP1b and pR8b, respectively, and for wild-type control strain ES5 containing pUC18NotI (Fig. 5A). The presence of pUC18NotI in the wild type reduced the number of bacteria recovered by approximately 0.1 log CFU/ml, which was statistically significant. For the two mutants, the presence of the empty vector did not lead to a further significant reduction in the number of bacteria recovered. For the complemented ESA_00282 mutant BF15(pR8b), the total number of cell-associated bacteria was the same as the number for the wild-type control containing pUC18NotI. For complemented mutant BF14(pP1b), the level was the same as the wild-type level.
FIG. 5.
Adhesion to Caco-2 intestinal epithelial cells. (A) Total cell-associated bacteria of wild-type strain ES5, strain ES5(pUC18NotI), mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn), and complemented mutants BF14 and BF15. One asterisk indicates that there is a significant difference from the results for wild-type strain ES5 (P < 0.01, Welch test), while two asterisks indicate that there is a significant difference from the results for strain ES5(pUC18NotI) (P < 0.001, Welch test). (B) Micrographs of Giemsa-stained Caco-2 monolayers with adherent bacteria (wild-type strain ES5 and mutants BF14 and BF15) taken at a magnification of ×1,000. Nuclei stained red-violet, and bacteria stained dark blue. Cl, cluster of bacteria; CP, cytoplasm; N, nucleus; S, single adherent bacterium.
The wild type adhered in clusters, and diffusive adhesion was visible between the clusters (Fig. 5B). For the flhE mutant (BF2) and the mgtB mutant (BF17), no differences from the wild type were observed. However, for mutants BF14 (ESA_00281::Tn) and BF15 (ESA_00282::Tn), clusters of bacteria were not present (Fig. 5B), and the occasional mutant clusters were much smaller than the wild-type clusters (not shown). Diffusively adherent bacteria appeared to be unchanged. Upon complementation, the wild-type pattern, with bacteria adhering both diffusively and in clusters, was observed for both mutants.
DISCUSSION
Cellulose biosynthesis mutants.
Bacterial cellulose biosynthesis is encoded in two operons, bcsABZC (yhjONML) and bcsEFG (yhjSTU), which are present in E. coli and Salmonella (9, 42, 52) and which were also recently characterized in C. sakazakii ES5 (15). Biofilm formation defects in cellulose biosynthesis operon mutants of Salmonella and E. coli have been reported previously (9, 42). Our screening also identified mutants defective in genes located in both operons, confirming once more that bcsG in the as-yet-uncharacterized bcsEFG operon has a role in cellulose biosynthesis and biofilm formation (42). For all of these mutants, the absence of cellulose from the extracellular matrix could be phenotypically confirmed on CR agar and calcofluor agar. The colony morphologies were similar to those described for cellulose operon mutants of Salmonella and E. coli (9, 52).
It is surprising that we did not detect any mutations in curli fimbria-associated genes, as the role of curli fimbriae in biofilm formation and as adhesins in both E. coli and Salmonella is well established (9, 38, 52). However, the structural genes for curli fimbriae are not present in the BAA-894 genome, and C. sakazakii wild-type strain ES5 formed rugose pink colonies on CR agar, a morphotype similar to the pink, dry, and rough (pdar) morphotype of Salmonella, which indicates the presence of cellulose but absence of curli in the extracellular matrix (38, 52). This finding implies that there are important differences from other Enterobacteriaceae in adhesion to abiotic as well as biotic surfaces and in biofilm formation.
Flagellar mutants.
Flagella seem to play a role in the initial phases of biofilm development, in attachment as well as further development (5). Our screening identified mutations in three flagellar genes, flgJ, fliD, and flhE.
EM investigations of the mutants defective in the flagellar muramidase FlgJ (mutant BF5) (34) and in the flagellar capping protein FliD (mutant BF2) (48) revealed defects that are consistent with previous reports for other species (26, 34).
FlhE is a flagellar protein whose function is currently unknown, and it is present in several proteobacterial genera (43). Cotranscription of flhE with the flagellar export apparatus genes flhAB as part of the flhABE operon has been confirmed for Salmonella enterica serovar Typhimurium (32, 43). FlhA and FlhB are required for flagellar biosynthesis (29) and, therefore, for both swimming and swarming motility. However, an flhE mutant of Salmonella serovar Typhimurium was defective only in swarming motility, and its flagellar structure was observed to be unaffected, which is in line with our EM results for the Cronobacter flhE mutant BF2 (43). Furthermore, the Salmonella serovar Typhimurium mutant also displayed other interesting phenotypes, such as enhanced biofilm formation on PVC, reduced calcofluor binding, and altered morphology on CR agar. Our results obtained with a C. sakazakii ES5 mutant, although not always in agreement with the phenotypes described for the corresponding Salmonella mutant, support the hypothesis of Stafford and Hughes that FlhE plays a role in the composition of the extracellular matrix (43).
Magnesium uptake and biofilm formation.
One mutant was found to be defective in a homologue of mgtB, which encodes a magnesium-transporting P-type ATPase. Although MgtB has been shown to mediate Mg2+ influx, at present it is not clear if this is its primary physiological function, as constitutive Mg2+ uptake in bacteria is usually mediated by CorA (41), which is also present in the BAA-894 genome (ORF ESA_03744). The fact that the mgtB homologue is a plasmid gene in BAA-894 further supports the notion that there is an ancillary magnesium uptake system. Interestingly, mgtE, encoding an Mg2+ channel, was found in a biofilm screening of Vibrio cholerae (33) and a screening of Aeromonas hydrophila for reduced adherence to Hep2-cells (31). The A. hydrophila mgtE mutant recovered also showed reduced adherence in a microtiter biofilm assay. Both V. cholerae and A. hydrophila possess CorA, so the mgtE product might also not be the main route of Mg2+ uptake in these organisms.
Genes with unknown functions.
Three mutants had disruptions in the uncharacterized ORFs ESA_00281 and ESA_00282, which are conserved in different Enterobacteriaceae but not in E. coli. In addition to the results of the CV microtiter assay, the corresponding mutants (BF14 and BF15, respectively) also had phenotypes different from the wild-type phenotype on CR agar, with respect to flow cell biofilm architecture, and with respect to adhesion to a biotic surface (see below). In each case, the phenotypes were the same for both mutant BF14 (ESA_00281::Tn) and mutant BF15 (ESA_00282::Tn), suggesting that the two genes are involved in the same process. Furthermore, all wild-type phenotypes could be restored at least partially by complementation. In Salmonella serovar Typhimurium ORFs STM3154 to STM3156 corresponding to ESA_00280 to ESA_00282 were described as motility genes (11, 45), and deficiencies in swimming and swarming were reported for mutants with mutations in these ORFs. The “microcolony-only” biofilm architecture observed for both mutant BF14 and mutant BF15 is consistent with this notion, as similar phenotypes have been reported for Pseudomonas aeruginosa motility mutant cultures, in which microcolonies arise through clonal growth of an immotile subpopulation, while the substratum surface is fully colonized by a motile subpopulation (5). In addition, the mutant colony morphology on CR agar also suggested a role in the composition of the extracellular matrix.
Adhesion of selected mutants to Caco-2 intestinal epithelial cells.
As some genetic factors involved in attachment to abiotic surfaces also play a role in adhesion to both plant and animal tissues (46, 49), we analyzed adhesion of selected mutants to Caco-2 cells. In these experiments, we observed that the absence of flagella (flgJ mutant BF13) greatly reduced the adhesion capacity, suggesting that flagella are important for Cronobacter adhesion to biotic surfaces, as has been shown for other Enterobacteriaceae (13, 37). An absence of cellulose had no effect on the adhesion capacity under our experimental conditions, which is in line with the results for Salmonella cellulose biosynthesis mutants reported by other workers (42). Also, mutants BF14 and BF15, which were defective in ORFs ESA_00281 and ESA_00282, respectively, in addition to having an altered adhesion pattern (only diffusively adherent bacteria compared with diffusively adherent bacteria and bacteria adhering in clusters), had significantly reduced adhesion capacities. However, the level of adhesion was still considerably higher than that of the aflagellate flgJ mutant BF13. As the Salmonella homologues of ESA_00281 and ESA_00282 were reported in the context of motility and flagella are known to be involved in surface and bacterial cell-to-cell adhesion, it could be assumed that these phenotypes are a consequence of dysfunctional or altered flagella. However, Cronobacter flhE mutant BF2, which is similar with respect to flagellum-related phenotypes (intact structure, swarming defects) to a Salmonella mutant (43), did not differ from the wild type in either adhesion capacity or adhesion pattern. Therefore, it is tempting to speculate that the mutations in ORFs ESA_00281 and ESA_00282 affect an adhesion factor(s) other than flagella that is involved in adhesion to and aggregation on Caco-2 cells.
In conclusion, this study indicated that two factors important for biofilm formation in other Enterobacteriaceae, cellulose and flagella, also contribute to biofilm formation in C. sakazakii. However, our results also revealed possible differences, such as the absence of curli fimbriae, at least in the isolate investigated in this study and strain C. sakazakii BAA-894, whose genome has been fully sequenced.
Acknowledgments
This work was supported by the Swiss National Science Foundation (project 3100A0-110039).
We thank Maya Grimm and Elisabeth Schraner for technical assistance, Susan Schönmann, Stefanie Heller, Chantal Loepfe, and Carmen Kaiser for experimental support, and Taurai Tasara and Peter Wild for helpful advice.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., J. C. Wootton, E. M. Gertz, R. Agarwala, A. Morgulis, A. A. Schaffer, and Y. K. Yu. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annous, B. A., P. M. Fratamico, and J. L. Smith. 2009. Scientific status summary. J. Food. Sci. 74:R24-37. [DOI] [PubMed] [Google Scholar]
- 5.Barken, K. B., S. J. Pamp, L. Yang, M. Gjermansen, J. J. Bertrand, M. Klausen, M. Givskov, C. B. Whitchurch, J. N. Engel, and T. Tolker-Nielsen. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331-2343. [DOI] [PubMed] [Google Scholar]
- 6.Bigot, S., V. Sivanathan, C. Possoz, F. X. Barre, and F. Cornet. 2007. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 64:1434-1441. [DOI] [PubMed] [Google Scholar]
- 7.Breeuwer, P., A. Lardeau, M. Peterz, and H. M. Joosten. 2003. Desiccation and heat tolerance of Enterobacter sakazakii. J. Appl. Microbiol. 95:967-973. [DOI] [PubMed] [Google Scholar]
- 8.Chenu, J. W., and J. M. Cox. 2009. Cronobacter (“Enterobacter sakazakii”): current status and future prospects. Lett. Appl. Microbiol. 49:153-159. [DOI] [PubMed] [Google Scholar]
- 9.Da Re, S., and J. M. Ghigo. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedemann, M. 2008. Enterobacter sakazakii in powdered infant formula. Bundesgesundheitsbl. Gesundheitsforsch. Gesundheitsschutz 51:664-674. (In German.) [DOI] [PubMed] [Google Scholar]
- 11.Frye, J., J. E. Karlinsey, H. R. Felise, B. Marzolf, N. Dowidar, M. McClelland, and K. T. Hughes. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 13.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 14.Glick, B. R. 1995. Metabolic load and heterologous gene expression. Biotechnol. Adv. 13:247-261. [DOI] [PubMed] [Google Scholar]
- 15.Grimm, M., R. Stephan, C. Iversen, G. G. Manzardo, T. Rattei, K. Riedel, A. Ruepp, D. Frishman, and A. Lehner. 2008. Cellulose as an extracellular matrix component present in Enterobacter sakazakii biofilms. J. Food. Prot. 71:13-18. [DOI] [PubMed] [Google Scholar]
- 16.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 17.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber, B., K. Riedel, M. Köthe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 19.Iversen, C., M. Lane, and S. J. Forsythe. 2004. The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38:378-382. [DOI] [PubMed] [Google Scholar]
- 20.Iversen, C., N. Mullane, B. McCardell, B. D. Tall, A. Lehner, S. Fanning, R. Stephan, and H. Joosten. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58:1442-1447. [DOI] [PubMed] [Google Scholar]
- 21.Johler, S., R. Stephan, I. Hartmann, K. A. Kuehner, and A. Lehner. 2010. Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl. Environ. Microbiol. 76:1053-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlyshev, A. V., M. J. Pallen, and B. W. Wren. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28:1078, 10810, 1082. [DOI] [PubMed] [Google Scholar]
- 23.Kim, H., J. Bang, L. R. Beuchat, and J. H. Ryu. 2008. Fate of Enterobacter sakazakii attached to or in biofilms on stainless steel upon exposure to various temperatures or relative humidities. J. Food. Prot. 71:940-945. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H., J. H. Ryu, and L. R. Beuchat. 2006. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 72:5846-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, H., J. H. Ryu, and L. R. Beuchat. 2007. Effectiveness of disinfectants in killing Enterobacter sakazakii in suspension, dried on the surface of stainless steel, and in a biofilm. Appl. Environ. Microbiol. 73:1256-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 28.Lehner, A., K. Riedel, L. Eberl, P. Breeuwer, B. Diep, and R. Stephan. 2005. Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J. Food. Prot. 68:2287-2294. [DOI] [PubMed] [Google Scholar]
- 29.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 30.Mange, J. P., R. Stephan, N. Borel, P. Wild, K. S. Kim, A. Pospischil, and A. Lehner. 2006. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merino, S., R. Gavin, M. Altarriba, L. Izquierdo, M. E. Maguire, and J. M. Tomas. 2001. The MgtE Mg2+ transport protein is involved in Aeromonas hydrophila adherence. FEMS Microbiol. Lett. 198:189-195. [DOI] [PubMed] [Google Scholar]
- 32.Minamino, T., T. Iino, and K. Kutuskake. 1994. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 176:7630-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller, R. S., D. McDougald, D. Cusumano, N. Sodhi, S. Kjelleberg, F. Azam, and D. H. Bartlett. 2007. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J. Bacteriol. 189:5348-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nambu, T., T. Minamino, R. M. Macnab, and K. Kutsukake. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181:1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuhäuser, M., and G. D. Ruxton. 2009. Distribution-free two-sample comparisons in the case of heterogeneous variances. Behav. Ecol. Sociobiol. 63:617-623. [Google Scholar]
- 36.Pang, S. S., R. G. Duggleby, R. L. Schowen, and L. W. Guddat. 2004. The crystal structures of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. J. Biol. Chem. 279:2242-2253. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, J. M., G. Grant, E. Allen-Vercoe, M. J. Woodward, A. Pusztai, and H. J. Flint. 2000. Adhesion of Salmonella enterica var. Enteritidis strains lacking fimbriae and flagella to rat ileal explants cultured at the air interface or submerged in tissue culture medium. J. Med. Microbiol. 49:691-696. [DOI] [PubMed] [Google Scholar]
- 38.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 39.Saiki, K., H. Nakamura, T. Mogi, and Y. Anraku. 1996. Probing a role of subunit IV of the Escherichia coli bo-type ubiquinol oxidase by deletion and cross-linking analyses. J. Biol. Chem. 271:15336-15340. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Smith, R. L., and M. E. Maguire. 1998. Microbial magnesium transport: unusual transporters searching for identity. Mol. Microbiol. 28:217-226. [DOI] [PubMed] [Google Scholar]
- 42.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 43.Stafford, G. P., and C. Hughes. 2007. Salmonella typhimurium flhE, a conserved flagellar regulon gene required for swarming. Microbiology 153:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao, L., R. E. Jackson, P. E. Rouvière, and Q. Cheng. 2005. Isolation of chromosomal mutations that affect carotenoid production in Escherichia coli: mutations alter copy number of ColE1-type plasmids. FEMS Microbiol. Lett. 243:227-233. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Q., S. Mariconda, A. Suzuki, M. McClelland, and R. M. Harshey. 2006. Uncovering a large set of genes that affect surface motility in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7981-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X., M. Rochon, A. Lamprokostopoulou, H. Lunsdorf, M. Nimtz, and U. Römling. 2006. Impact of biofilm matrix components on interaction of commensal Escherichia coli with the gastrointestinal cell line HT-29. Cell. Mol. Life Sci. 63:2352-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild, P. 2008. Electron microscopy of viruses and virus-cell interactions. Methods Cell Biol. 88:497-524. [DOI] [PubMed] [Google Scholar]
- 48.Yonekura, K., S. Maki, D. G. Morgan, D. J. DeRosier, F. Vonderviszt, K. Imada, and K. Namba. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148-2152. [DOI] [PubMed] [Google Scholar]
- 49.Yousef-Coronado, F., M. L. Travieso, and M. Espinosa-Urgel. 2008. Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol. Lett. 288:118-124. [DOI] [PubMed] [Google Scholar]
- 50.Yu, C. S., Y. C. Chen, C. H. Lu, and J. K. Hwang. 2006. Prediction of protein subcellular localization. Proteins 64:643-651. [DOI] [PubMed] [Google Scholar]
- 51.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]