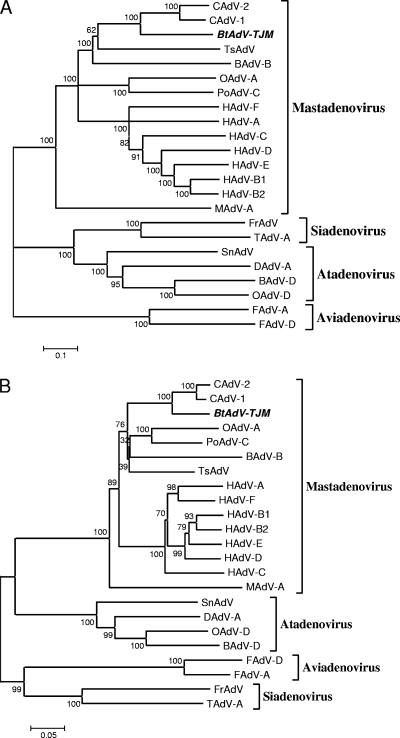
FIG. 4.
Phylogenetic analysis of bat adenoviruses. The phylogenetic trees were constructed based on the alignment of full-length AdV genome nucleotide sequences (A), the amino acid sequences of hexon proteins (B), and partial amino acid sequences of the polymerases (C) of bat adenoviruses by using the neighbor-joining (NJ) method with a bootstrap of 1,000 replicates with the MEGA 4.1 program. Gaps were regarded as a complete deletion unless otherwise specified. A condensed tree is presented in C. The scale bar is in units of nucleotide (A) or amino acid (B and C) residue substitutions per site. Bootstrap values (percent) are indicated at the branch nodes of the phylogenetic trees. Abbreviations are as follows (with GenBank accession numbers in parentheses): HAdV-A, human adenovirus A (accession no. NC_001460); HAdV-B1, human adenovirus B1 (accession no. NC_011203); HAdV-B2, human adenovirus B2 (accession no. NC_011202); HAdV-C, Human adenovirus C (NC_001405); HAdV-D, human adenovirus D (accession no. NC_010956); HAdV-E, human adenovirus E (accession no. NC_003266); HAdV-F, human adenovirus F (accession no. NC_001454); OAdV-A, ovine adenovirus A (accession no. NC_002513); OAdV-D, ovine adenovirus D (accession no. NC_004037); PoAdV-C, porcine adenovirus C (accession no. NC_002702); BAdV-B, bovine adenovirus B (accession no. NC_001876); BAdV-D, bovine adenovirus D (accession no. NC_002685); CAdV-1, canine adenovirus 1 (accession no. NC_001734); CAdV-2, canine adenovirus 2 (accession no. AC_000020); MAdV-A, murine adeno- virus A (accession no. NC_000942); TsAdV, tree shrew adenovirus (accession no. NC_004453); SnAdV, snake adenovirus (accession no. NC_009989); FrAdV, frog adenovirus (accession no. NC_002501); TAdV-A, turkey adenovirus A (accession no. NC_001958); FAdV-A, fowl adenovirus A (accession no. NC_001720); FAdV-D, fowl adenovirus D (accession no. NC_000899); DAdV-A, duck adenovirus A (accession no. NC_001813); bat AdV-FBV1, bat adenovirus FBV1 (accession no. AB303301). The bat AdV detected in this study is named by the sample number followed by the abbreviation of the sampling location and species name. The sequences derived from different locations are highlighted with different symbols. HB, Hubei; HN, Hainan; GD, Guangdong; YN, Yunnan; TJ, Tianjin; Ha, Hipposideros armiger; Md, Myotis daubentoni; Mh, Myotis horsfieldii; Mspp, unidentified Myotis species; Mr, Myotis ricketti; SK, Scotophilus kuhlii.