Abstract
Chronic infection with hepatitis B virus (HBV) is an important cause of cirrhosis and cancer of the liver. HBV is currently classified into eight genotypes, A to H. Accumulated evidence shows that the genotype influences both the clinical course of infection and the response to treatment. We describe a new method for genotyping based on TaqMan real-time PCR, which identifies all HBV genotypes without post-PCR processing. In this assay, each sample is processed in four multiplex real-time PCRs, each targeting two or three genotype-specific segments of HBV. By analyzing 185 samples representing all genotypes and different proportions of genotype mixtures, we could validate high accuracy of the assay. We conclude that this new assay represents a significant advancement for both diagnostics and clinical research because it is accurate, practical, and based on a technique that is well established in many virological laboratories.
Hepatitis B virus (HBV) has been classified into eight genotypes, A to H, some of which have been further divided into a number of subgenotypes. The original geographical distribution of the genotypes is well known (17, 24), but as a result of migration and traveling, the predominant genotypes are today A and D in Europe; A, D, and E in Africa; D in Central Asia and the Middle East; A, C, and D in India; B and C in East Asia; and A, D, F, and H in South America. Essentially all genotypes are prevalent in multiethnic societies such as in North America and Australia (6, 28).
During the past decade, accumulating evidence has demonstrated that the HBV genotype has an impact on both prognosis and treatment response, as well as on the pattern of mutations emerging in the viral DNA (29). In particular, patients with genotype C are expected to experience a poorer response to treatment and a worse prognosis in comparison with those infected with genotype B, and genotype A patients more frequently experience normalization of alanine aminotransferase (ALT) levels, clearance of viral DNA, and clearance of HBsAg than genotype D patients (16, 27). Due to the clinical importance of the infecting genotype, it is likely that genotyping of hepatitis B will become increasingly requested in the clinical appraisal and before treatment of infected individuals.
Several genotyping methods have been described, based mainly on sequencing, restriction fragment length polymorphism (RFLP), PCR with genotype-specific primers, line probe assay, or real-time PCR (4, 15, 17, 18, 20, 23, 25). Some of these assays have limited accuracy, and most of them are suboptimal for modern high-throughput molecular diagnostics. Here we describe a method that makes use of the TaqMan real-time PCR in a multiplex manner in order to identify all genotypes without the need for post-PCR steps. The assay is easy to use and therefore is well suited for routine application in the modern diagnostic laboratory.
MATERIALS AND METHODS
Samples.
A total of 184 clinical samples representative of genotypes A through G were included, of which 129 had been previously genotyped by RFLP (17). The remaining 55 were analyzed by RFLP and/or by sequencing in parallel with the TaqMan genotyping. The genotype G sample was kindly provided by Stephan Günther (Hamburg, Germany).
As no sample representing genotype H was available, a synthetic control was constructed, carrying a segment of the pre-S/S region of HBV genotype H inserted into a pUC57 plasmid. This plasmid was synthesized by GenScript Corporation (Piscataway, NJ).
DNA extraction and postelution.
Viral DNA was extracted from samples by a Magnapure LC robot (Roche Applied Science, Mannheim, Germany) with the DNA I protocol. In a postelution step, also performed by the Magnapure robot, the extracted DNA was loaded into a 96-well PCR plate that was preloaded with the four genotyping reaction mixtures (as described below).
Multiplex TaqMan PCR.
The MacVector software (MacVector Incorporated, Cary, NC) was used for detailed investigation of the aligned sequences retrieved from databases and representative of all genotypes. This analysis aimed at identifying signature nucleotides characteristic of each genotype. These positions were then targeted by minor groove binding (MGB) probes or by the 3′ end of the forward or reverse primer in order to achieve discrimination. The Primer Express (Applied Biosystems, Foster City, CA) software was used to obtain suitable annealing temperatures for the primers and probes.
Ten sets of forward and reverse primers and 13 MGB probes were combined in four multiplex reaction mixtures (Table 1; Fig. 1). Thus, each genotype was identified by one or two specific real-time amplification sets, using either primer or probe discrimination or a combination of both (Table 1). For each of genotypes A, B, and C, two sets of primers and probes (AS/AC, BS/BpS, and CS/CpS) were designed in order to correctly identify the various subgenotypes (21, 26, 32) as well as strains with untypical sequences (Fig. 2). Moreover, both probes of the A sets were complemented with one additional probe each, for a satisfactory coverage of divergence in the target regions. The two resulting mixtures of probes were simulations of two single probes with degenerate bases. The designations in subscript of the dual sets of genotypes A through C indicate the region (S, core, and pre-S) of the start position of the forward primer in each set.
TABLE 1.
Summary of primers and probes used in TaqMan PCR genotyping analysis
| Multiplex reaction mixture and designation | Primer or probea | Genotype | Detectorb | Sense/antisense | Key oligonucleotidec | Sequence (5′→3′)d |
|---|---|---|---|---|---|---|
| 1 | ||||||
| ASe | Forward | A | S | × | CATCTTCTTRTTGGTWCTTCTGGAT | |
| Reverse | AS | GCAKGGTCCCGTRCTGGTT | ||||
| MGB probe | FAM | S | CTCTAATTCCAGGATCMACA | |||
| BpS | Forward | B | VIC | S | × | GCATGGGGACAAATCTTTCTGTC |
| Reverse | AS | × | AATCTGGATTKTCTGAGTTGGCTTT | |||
| MGB probe | S | × | CCCTGGGATTCTTC | |||
| CpS | Forward | C | S | × | TGCACCGAACATGGAGATCACf | |
| Reverse | AS | TCTGTGGTATTGTGAGGATTCTTGTC | ||||
| MGB probe | NED | S | ACCCCTGCTCGTGTTA | |||
| 2 | ||||||
| E | Forward | E | S | CCTCATTTTGTGGGTCACCWTATTC | ||
| Reverse | AS | × | CCATTCGAGAGGGACCGTC | |||
| MGB probe | VIC | AS | × | AGCCCCATGATGTAGC | ||
| FHg | Forward | F and H | S | × | CCGACTATTGCCTCTCTCACATCA | |
| Reverse | AS | GGGGTCCTAGGAGTCCTGATGT | ||||
| MGB probe | NED | S | × | CCCTGCTATGAACATGGA | ||
| G | Forward | G | S | × | GAAACCGCCATGAACACCTCT | |
| Reverse | AS | × | CCGGTTGTTGACATAACAAACAGT | |||
| MGB probe | FAM | S | × | TCTGCCAAGGCAGTTAT | ||
| 3 | ||||||
| ACe | Forward | A | S | AAATGCCCCTATCTTATCAACACTTC | ||
| Reverse | AS | TGCGAGGCGAGGGAGTTCT | ||||
| MGB probe | FAM | AS | × | CTCKGTCYCGTCGTCTAA | ||
| BS | Forward | B | S | AGACTCGTGGTGGACTTCTCTCA | ||
| Reverse | AS | × | CCAGGACAAATTGGAGGACAAC | |||
| MGB probe | VIC | S | × | CCAAATCTCCAGTCACTC | ||
| Qh | MGB probe | A through F | NED | S | NAi | CAGGTCCCCTAGWAGA |
| 4 | ||||||
| CS | Forward | C | S | GTATGTTGCCCGTTTGTCCTCTAC | ||
| Reverse | AS | GGARTCGTGCAGGTCTTGCA | ||||
| MGB probe | NED | S | × | CAGGAACATCAACTACCAGC | ||
| D | Forward | D | S | CTCATTTTGTGGGTCACCATATTC | ||
| Reverse | AS | GGTCGGGAAAGAATCCCAGA | ||||
| MGB probe | FAM | S | × | CAGAATCTTTCCACCAGCA |
MGB, minor groove binding moiety.
FAM, 6-carboxyfluorescein; VIC and NED, fluorophores proprietary to Applied Biosystems (ABI).
Markings indicate whether the sets were designed using primer or probe specificity or a combination.
Marked positions discriminate between genotypes: boldface nucleotides are unique for the genotype, whereas underlined nucleotides individually mismatch against at least three other genotypes.
Both genotype A sets are equipped with two MGB probes each for a satisfactory coverage of the divergences in the regions of interest. The total probe concentration in each of the two sets is equal to that of all the other sets. The four probes are depicted as two with degenerate bases.
Italic denotes a mismatch introduced to improve the performance of the CpS set.
The FH set in multiplex reaction mixture 2 hybridizes to both genotypes F and H. In case of a positive result in the first analysis, the sample has to be reanalyzed in a supplementary run to distinguish between the two genotypes, using differential reverse primers gtF_R and gtH_R in two singleplex mixtures, as described in Materials and Methods.
The Q probe works with primers included in the AC set and was designed to hybridize to all genotypes except G and H.
NA, not applicable.
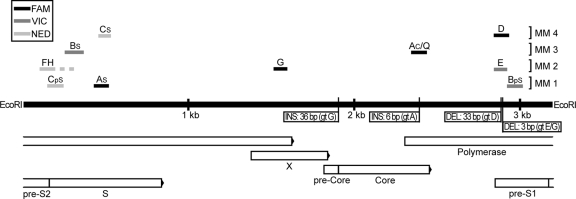
FIG. 1.
Schematic diagram of the hepatitis B genome and the genotyping system. Genes are depicted as horizontal bars below the black line representing the genome. Vertical lines indicate insertions and deletions in the genome. Horizontal lines above the genome represent amplicons produced by the genotyping sets. The broken line expanding from the FH set shows the longer amplicon resulting from supplementary analysis used for differentiation of genotypes F and H. EcoRI, unique cleavage site for EcoRI enzyme; INS, insertion; DEL, deletion; MM, multiplex reaction mixture; FAM, 6-carboxyfluorescein; VIC and NED, fluorophores proprietary to Applied Biosystems (ABI).
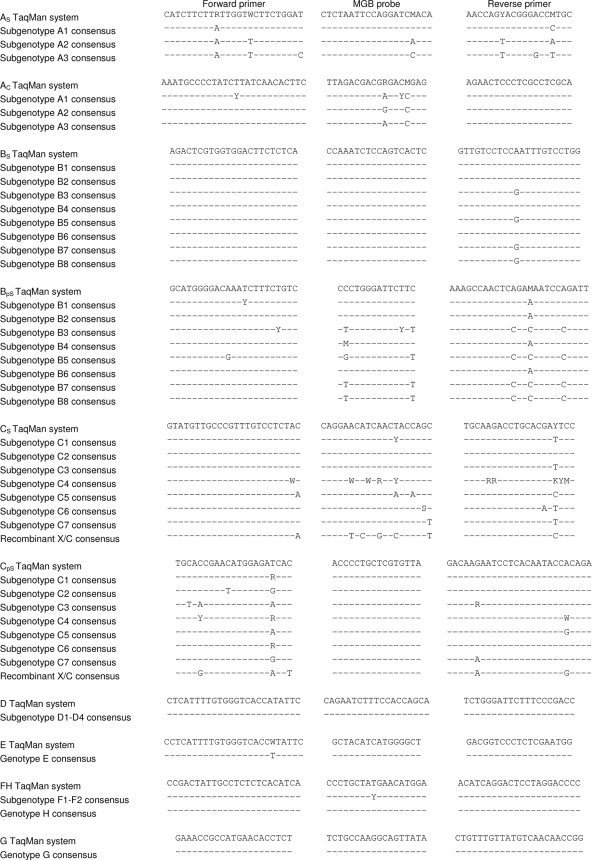
FIG. 2.
Sequences for the forward primer, MGB probe, and reverse primer, respectively, in each set, in comparison with the corresponding consensus sequences of genotypes and subgenotypes of interest. Only positions in the viral DNA differing from the TaqMan primer and probe sequences are shown. The X/C recombinant was first described by Hannoun et al. (9).
Multiplex mixtures 1 and 2 identify genotypes A through C and E through H, respectively, whereas mixtures 3 and 4 identify genotypes A and B and genotypes C and D, respectively. The forward primer in the CpS set, which anneals only to genotype C, was designed with one additional mismatch four nucleotides from the 3′ end, to improve performance and discrimination (Fig. 2). The FH set was designed to identify both genotypes F and H. Where a positive reaction showed genotype F or H, the sample was reanalyzed by using specific reverse primers to distinguish genotype. These primers (HBVgtF_R [GCCAGGACACCCGGGTAKTC] and HBVgtH_R [CCAGGACACCCGGGTGGTA]) were used in combination with the original FH forward primer and MGB probe in two singleplex reactions. The third multiplex mixture was equipped with a control probe, Q, with the ability to hybridize to amplicons of all genotypes except G and H, in the reaction initiated by the AC primers. This Q probe was included in order to detect atypical strains that might not be identified by any of the genotype-specific systems. Moreover, the Q probe also served as a quantitative estimate of the HBV DNA level and to give information in advance about a potential genotype F or H sample, as the ability of the Q probe to hybridize to a genotype H sample is limited.
The genotyping analysis was performed on an ABI 7300 real-time PCR instrument (Applied Biosystems, Foster City, CA). Each sample required four wells, and hence the maximum capacity was 24 samples, including one or several controls per PCR plate. The 50-μl reaction volume contained 25 μl Universal PCR master mix (Roche Diagnostics, Branchburg, NJ), 0.2 μM each primer and probe, and 10 μl of extracted DNA. After uracil DNA glycosylase activation at 50°C for 2 min and initial denaturation at 95°C for 10 min, the PCR was run for 45 cycles with denaturation at 95°C for 15 s and a combined step of annealing and extension at 60°C for 1 min. The same concentrations and temperatures were valid also for the two supplementary singleplex mixtures used for distinguishing between genotypes F and H.
Sensitivity.
The sensitivity was evaluated by serial dilutions, in 1:10 steps, of 10 clinical samples representing genotypes A through E. The HBV DNA levels had previously been quantified with the Cobas TaqMan assay (Roche Diagnostics, Branchburg, NJ).
Genotype mixtures.
Clinical samples representing genotypes A, B, C, D, and G, with HBV DNA levels identified by Cobas TaqMan, were tested in different combinations (A/D, A/G, and B/C) with different proportions to simulate mixed genotypes. All mixtures were analyzed in duplicate.
RESULTS
Analytical performance.
The amplification efficacy for each component PCR, examined by testing serial dilutions of 10 clinical samples representing various genotypes, was in the range of 85 to 93%. The lowest detected levels among these diluted samples were 1.5 to 3 log IU/ml (with the lower value valid for genotypes A, B, and D). The clinical samples used for evaluation of this method had viral concentrations of between 2.5 log IU/ml and 9.0 log IU/ml, yielding threshold cycle (CT) values of 12.1 through 41.0.
Clinical performance.
As can be seen in Table 2, there was good concordance between the TaqMan method and genotyping by sequencing or RFLP. The same genotype was identified in 173 samples, including 23 A, 26 B, 35 C, 77 D, 7 E, 2 F, and 1 G. A mixture of genotypes A and D was observed in one sample previously analyzed as genotype D only. One genotype E sample and three samples of subgenotype C5 were reactive only by the Q probe. The six samples harboring an X/C recombinant, recently proposed to be classified as a new genotype designated I (9, 10), produced results indicating a difficult-to-type strain: four samples produced only a Q-probe reactivity, and two were weakly reactive with the AS set in addition to having a Q-probe reactivity. Two samples (1.1%) had a low viral concentration and were negative by the TaqMan genotyping PCR.
TABLE 2.
Results of HBV genotyping of 185 samples also genotyped by sequencing or RFLP
| Sequencing/RFLP genotype (no. of samples)a | No. (%) of samples with TaqMan genotypeb: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | Only Q probe | Nondetectable | Mixed A/D | |
| A (24) | 23 (96) | 1 | |||||||||
| B (26) | 26 (100) | ||||||||||
| C (38) | 35 (92) | 3c | |||||||||
| D (79) | 77 (97) | 1 | 1 | ||||||||
| E (8) | 7 (88) | 1 | |||||||||
| F (2) | 2 (100) | ||||||||||
| G (1) | 1 (100) | ||||||||||
| Hd (1) | 1 (100) | ||||||||||
| X/C recombinante (6) | 6f | ||||||||||
A total of 148 samples were identified by RFLP (19 of them were also sequenced, with congruent results), and 36 were genotyped only by sequencing.
Numbers in parentheses are percentages of the corresponding number of samples genotyped by sequencing or RFLP.
Subgenotype C5 strains (by sequencing).
Genotype H was represented by a pUC57 plasmid carrying a synthetic segment of the pre-S/S region of HBV genotype H.
The X/C recombinant strain, first described by Hannoun et al. (9), is made up partly by genotype C DNA and partly by untypeable DNA.
Two of these six samples also showed weak reactivity with the AS set, but with CT values >10 cycles higher than for Q.
Testing genotype mixtures.
The genotype mixtures were prepared and analyzed in five different proportions (0:100, 20:80, 50:50, 80:20, and 100:0) to evaluate the possibility of demonstrating coinfection in samples from patients. Genotype A was mixed with D and G and genotype B with C (Table 3). The A-D mixture showed high conformity in all related PCR sets, without any cross-reactivity. The shift in proportions between genotypes A and D were well reflected by the reciprocal change in CT values, decreasing from 27.0 to 24.8 for A and increasing from 25.5 to 27.7 for D. CT values for the Q probe did not change, as expected since the total viral concentration remained at the same level. The A-G mixture showed similar results with the two A sets, with changes in CT values as the proportion of genotypes A and G shifted. Because the Q probe cannot hybridize to genotype G, the CT values of the Q probe were not stable and changed with the concentration of the genotype A component of the mixture. The 50:50 mixture showed a difference in CT value of about 2.5 cycles between genotypes A and G, corresponding to a 5.6-fold difference in viral concentration. This discrepancy was conserved in all proportions and was probably due to the fact that the genotype G sample could not be calibrated using the Q probe, as was done with the other samples before analysis. In the B-C mixture, the CT for the Q set was stable, as in the A-D mixture. There were some small disparities between the two B sets and the two C sets. CT values for BS were about 2.5 cycles later than those for BpS, whereas CT values for CS were about 1.5 cycles later than those for CpS. Thus, for the samples used in this mixture, the concentrations were better described by the CT values from the BpS and CpS sets than by those from the BS and CS sets, with a difference of 2.5 to 3 cycles between those pairs in the 50:50 mixture.
TABLE 3.
CT values resulting from the analysis of simulated genotypic mixtures
| Genotype mixture and proportion |
CT valuea |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AS | BpS | CpS | E | FH | G | AC | BS | Q | CS | D | |
| A-D | |||||||||||
| 0:100 | 27.1 | 25.5 | |||||||||
| 20:80 | 27.0 | 27.0 | 27.2 | 25.8 | |||||||
| 50:50 | 25.8 | 25.6 | 26.5 | 26.2 | |||||||
| 80:20 | 25.2 | 25.0 | 26.5 | 27.7 | |||||||
| 100:0 | 24.9 | 24.8 | 26.5 | ||||||||
| A-G | |||||||||||
| 0:100 | 27.2 | ||||||||||
| 20:80 | 27.3 | 27.6 | 26.9 | 28.9 | |||||||
| 50:50 | 25.8 | 28.2 | 25.7 | 27.2 | |||||||
| 80:20 | 25.1 | 29.7 | 24.9 | 26.9 | |||||||
| 100:0 | 24.5 | 24.3 | 26.5 | ||||||||
| B-C | |||||||||||
| 0:100 | 28.0 | 27.3 | 29.4 | ||||||||
| 20:80 | 29.7 | 28.5 | 34.2 | 27.4 | 30.1 | ||||||
| 50:50 | 28.7 | 28.8 | 31.9 | 27.0 | 31.1 | ||||||
| 80:20 | 28.2 | 32.0 | 30.9 | 26.7 | 32.1 | ||||||
| 100:0 | 28.1 | 30.6 | 27.2 | ||||||||
All mixtures were run in duplicate; mean CT values are shown.
DISCUSSION
The aim of this study was to develop and evaluate a modern genotyping method that would allow specific determination and discrimination of all known HBV genotypes. The method was designed for genotyping, and should therefore be utilized in complement to an assay for detection and quantification. We chose a method based on real-time PCR with hydrolysis (TaqMan) probes, because this technique is now well established in many diagnostic laboratories. The TaqMan technique is suitable for typing, because specificity can be obtained by differences in either probe matching (≥2 mismatches preclude proper probe hybridization) or in primer 3′ matching (≥1 mismatch impairs primer elongation and thus amplification). Moreover, real-time PCR has the general advantage of technical simplicity without post-PCR steps and a low risk of false results due to contamination. It was not possible, however, to find one single genomic segment that could be used for amplification and identification of all genotypes. Instead, we chose to target a number of segments in different parts of the genome to obtain reliable typing accuracy (Fig. 1).
In the final version, genotypes D through H could be detected by one single amplification set for each genotype. For the genotypes A, B, and C, it was more difficult to find targets that were sufficiently conserved and not liable to erroneous results due to single point mutations. Therefore, we chose to apply two amplification sets for each of genotypes A, B, and C. Figure 2 lists all genotype-specific oligonucleotides and the corresponding targets in the viral genome. Both genotype A sets detect subgenotypes A1 and A2, whereas A3 is predicted to be identified only by the AC set, mainly due to a single mismatch in the 3′ end of the AS set forward primer against this subgenotype. For genotype B, the BS set can distinguish all eight subgenotypes, whereas the BpS set is predicted to detect only B1, B2, B4, and B6. Due to somewhat more irregular differences among the C subgenotypes, the CS and CpS sets function in an even more complementary manner. Hence, subgenotypes C1 and C2, most strains of C6 and C7, and some strains of C3 and C4 should be detected by both genotype C sets. Because we found that subgenotype C5 and the X/C recombinant were detected only by the Q probe, we designed specific TaqMan systems also for these variants (details are provided in the supplemental material). For genotypes A, B, and C, subgenotypes with mismatches often produce CT values from both sets but with a difference of about 5 to 10 cycles. In summary, nearly all currently known subgenotypes of hepatitis B virus are efficiently covered by this method.
The accuracy of this new genotyping method was evaluated by analyzing 185 samples in which the genotype had previously been assessed by RFLP or sequencing. The genotype was correctly identified in 173 samples with the new assay. Two samples were undetectable, probably due to a low HBV DNA level. Ten samples could not be genotyped but produced amplicons detected by the Q probe, indicating the presence of HBV DNA that might represent a divergent strain. Indeed, six of them contained a recombinant HBV strain, which recently was proposed to be classified as a new genotype, I (9, 10). The four remaining untypeable samples were of genotype C5 and E. Overall, these results yield a sensitivity of 94% and a specificity of 100%.
In one sample classified as genotype D, the TaqMan assay identified a coinfection with genotype A in addition to the previously identified genotype D. This is one example of the strength of real-time PCR, by which each genotype can be specifically amplified and/or detected. The occurrence of genotypic mixtures in hepatitis B infections, especially among patients on treatment, has been reported by several groups. Generally, the coinfecting minor strain is masked by the major one and is therefore scarcely detected with methods such as sequencing (3, 8, 11). To evaluate the ability of the TaqMan assay to identify genotype coinfections, we analyzed mixtures of genotypes with various genotype proportions. We found that a coinfecting strain, constituting 10 to 20% or less of the total viremia, could be clearly identified. The approximate size of the proportions could be estimated by comparing the CT values for the respective genotypes. However, such estimates should be done with caution because the amplification efficiencies of the various component PCRs may differ.
Some of the component PCRs used primers that amplified several or all genotypes, and in these cases discrimination relied only upon probe mismatches. In such cases a certain degree of cross-reactivity was observed, but this was possible to distinguish from coinfection because the amplification curves were flat and appeared later, yielding CT values that were typically more than 10 cycles higher than the CT value for the correct genotype.
It is not yet well established how genotyping should be used in clinical diagnostics despite accumulating evidence for clinically important differences between genotypes. Further clinical studies on the genotypic influence on hepatitis B virus infection (7, 22, 29), especially in the United States, Europe, and Africa (on genotypes A and D through H), are therefore needed, because most studies to date have been performed in Asia, where genotypes B and C prevail. Genotype C has been shown to induce more severe chronic inflammation of the liver than genotype B and thus is associated with a higher risk of developing liver cirrhosis and hepatocellular carcinoma (HCC) (2, 5, 12, 19). Genotype C has even been identified as an independent risk factor for the development of HCC (1). Furthermore, genotypes A and B have been reported to be associated with an earlier seroconversion from HBeAg to anti-HBe than genotypes C and D (5, 16). However, in a study from South Africa, subgenotype A1 was associated with a higher risk for HCC than other subgenotypes of A or non-A genotypes (14). Genotype impact on treatment outcome is more uncertain, but genotype B has been shown to respond better to alpha interferon treatment than genotype C (13, 30). As for nucleoside analogues, the effect of lamivudine but not that of adefovir appears to be affected by the infecting genotype (31). These observations have not yet been translated into strict guidelines, but they suggest that genotyping should be part of the clinical evaluation of patients, because knowledge of the genotype may help to predict the course of infection and the choice of treatment. Indeed, an increasing demand for HBV genotyping has already been noted at our laboratory. To meet this demand there is a need for a rapid and easy-to-use genotyping method suitable for a modern diagnostic laboratory. Such a method should be capable of identifying all genotypes, and this is of particular importance in multiethnic societies, where all genotypes may appear as a result of human migration. We chose to develop a method based on TaqMan PCR because in our clinical diagnostics all other viruses are identified by this technology. The method described here identifies all eight genotypes, A through H, and avoids several of the laborious and time-consuming procedures that are associated with some of the older methods, since the analysis is made without any post-PCR steps. Twenty-four samples (one 96-well plate) require about 4 h to be processed, including DNA extraction, postelution, and TaqMan PCR. In combination with an in-house spreadsheet template document (available upon request) used as a programming interface and a link between the instruments, the viral DNA is easily analyzed in a straightforward manner, without unnecessary delay.
An advantage with the real-time PCR genotyping is that it produces CT values for the identified genotypes that give an indication of the HBV DNA concentration. By analyzing mixtures of genotypes, we could show that mixed infection was accurately identified by clear-cut signals for each genotype and also that the relative proportion of each genotype was well represented by its CT value.
To conclude, by targeting multiple segments of the genome we have developed a real-time PCR assay that can identify all HBV genotypes in a one-step procedure. In addition to its simplicity and genotyping accuracy, the method also may give an indication of HBV DNA concentration and, in case of mixed infections, of the relative proportions of several genotypes.
Supplementary Material
Acknowledgments
This work was financially supported by the ALF Funds (grant ALFGBG-2983).
We thank Stephan Günther for the kind provision of a genotype G sample and Anne-Sofie Tylö for technical expertise.
Footnotes
Published ahead of print on 27 January 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Chan, H. L., A. Y. Hui, M. L. Wong, A. M. Tse, L. C. Hung, V. W. Wong, and J. J. Sung. 2004. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut 53:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, H. L., M. L. Wong, A. Y. Hui, L. C. Hung, F. K. Chan, and J. J. Sung. 2003. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J. Clin. Microbiol. 41:1277-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, B. F., P. J. Chen, G. M. Jow, E. Sablon, C. J. Liu, D. S. Chen, and J. H. Kao. 2004. High prevalence of mixed genotype infections in hepatitis B virus infected intravenous drug users. J. Med. Virol. 74:536-542. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., J. Yin, X. Tan, H. Zhang, H. Zhang, B. Chen, W. Chang, S. Schaefer, and G. Cao. 2007. Improved multiplex-PCR to identify hepatitis B virus genotypes A-F and subgenotypes B1, B2, C1 and C2. J. Clin. Virol. 38:238-243. [DOI] [PubMed] [Google Scholar]
- 5.Chu, C. J., M. Hussain, and A. S. Lok. 2002. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 122:1756-1762. [DOI] [PubMed] [Google Scholar]
- 6.Chu, C. J., E. B. Keeffe, S. H. Han, R. P. Perrillo, A. D. Min, C. Soldevila-Pico, W. Carey, R. S. Brown, Jr., V. A. Luketic, N. Terrault, and A. S. Lok. 2003. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology 125:444-451. [DOI] [PubMed] [Google Scholar]
- 7.Chu, C. J., and A. S. Lok. 2002. Clinical significance of hepatitis B virus genotypes. Hepatology 35:1274-1276. [DOI] [PubMed] [Google Scholar]
- 8.Hannoun, C., K. Krogsgaard, P. Horal, and M. Lindh. 2002. Genotype mixtures of hepatitis B virus in patients treated with interferon. J. Infect. Dis. 186:752-759. [DOI] [PubMed] [Google Scholar]
- 9.Hannoun, C., H. Norder, and M. Lindh. 2000. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J. Gen. Virol. 81:2267-2272. [DOI] [PubMed] [Google Scholar]
- 10.Huy, T. T. T., T. N. Trinh, and K. Abe. 2008. New complex recombinant genotype of hepatitis B virus in Vietnam. J. Virol. 82:5657-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jardi, R., F. Rodriguez-Frias, M. Schaper, E. Giggi, D. Tabernero, M. Homs, R. Esteban, and M. Buti. 2008. Analysis of hepatitis B genotype changes in chronic hepatitis B infection: influence of antiviral therapy. J. Hepatol. 49:695-701. [DOI] [PubMed] [Google Scholar]
- 12.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 13.Kao, J. H., N. H. Wu, P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes and the response to interferon therapy. J. Hepatol. 33:998-1002. [DOI] [PubMed] [Google Scholar]
- 14.Kew, M. C., A. Kramvis, M. C. Yu, K. Arakawa, and J. Hodkinson. 2005. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-Saharan Africans. J. Med. Virol. 75:513-521. [DOI] [PubMed] [Google Scholar]
- 15.Kirschberg, O., C. Schuttler, R. Repp, and S. Schaefer. 2004. A multiplex-PCR to identify hepatitis B virus-genotypes A-F. J. Clin. Virol. 29:39-43. [DOI] [PubMed] [Google Scholar]
- 16.Lin, C. L., and J. H. Kao. 2008. Hepatitis B viral factors and clinical outcomes of chronic hepatitis B. J. Biomed Sci. 15:137-145. [DOI] [PubMed] [Google Scholar]
- 17.Lindh, M., A. S. Andersson, and A. Gusdal. 1997. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 18.Lindh, M., J. E. Gonzalez, G. Norkrans, and P. Horal. 1998. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J. Virol. Methods 72:163-174. [DOI] [PubMed] [Google Scholar]
- 19.Lindh, M., P. Horal, A. P. Dhillon, and G. Norkrans. 2000. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J. Viral Hepat. 7:258-267. [DOI] [PubMed] [Google Scholar]
- 20.Liu, W. C., M. Mizokami, M. Buti, M. Lindh, K. C. Young, K. T. Sun, Y. C. Chi, H. H. Li, and T. T. Chang. 2006. Simultaneous quantification and genotyping of hepatitis B virus for genotypes A to G by real-time PCR and two-step melting curve analysis. J. Clin. Microbiol. 44:4491-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, W. C., P. H. Phiet, T. Y. Chiang, K. T. Sun, K. H. Hung, K. C. Young, I. C. Wu, P. N. Cheng, and T. T. Chang. 2007. Five subgenotypes of hepatitis B virus genotype B with distinct geographic and virological characteristics. Virus Res. 129:212-223. [DOI] [PubMed] [Google Scholar]
- 22.Madan, K., Y. Batra, V. Sreenivas, M. Mizokami, Y. Tanaka, S. B. Chalamalasetty, S. K. Panda, and S. K. Acharya. 2009. HBV genotypes in India: do they influence disease severity? Hepatol. Res. 39:157-163. [DOI] [PubMed] [Google Scholar]
- 23.Naito, H., S. Hayashi, and K. Abe. 2001. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J. Clin. Microbiol. 39:362-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norder, H., A. M. Courouce, P. Coursaget, J. M. Echevarria, S. D. Lee, I. K. Mushahwar, B. H. Robertson, S. Locarnini, and L. O. Magnius. 2004. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology 47:289-309. [DOI] [PubMed] [Google Scholar]
- 25.Osiowy, C., and E. Giles. 2003. Evaluation of the INNO-LiPA HBV genotyping assay for determination of hepatitis B virus genotype. J. Clin. Microbiol. 41:5473-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto, T., Y. Tanaka, E. Orito, J. Co, J. Clavio, F. Sugauchi, K. Ito, A. Ozasa, A. Quino, R. Ueda, J. Sollano, and M. Mizokami. 2006. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines. J. Gen. Virol. 87:1873-1882. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Tapias, J. M., J. Costa, A. Mas, M. Bruguera, and J. Rodes. 2002. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology 123:1848-1856. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer, S. 2007. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J. Gastroenterol. 13:14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer, S. 2005. Hepatitis B virus: significance of genotypes. J. Viral Hepat. 12:111-124. [DOI] [PubMed] [Google Scholar]
- 30.Wai, C. T., C. J. Chu, M. Hussain, and A. S. Lok. 2002. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology 36:1425-1430. [DOI] [PubMed] [Google Scholar]
- 31.Westland, C., W. T. Delaney, H. Yang, S. S. Chen, P. Marcellin, S. Hadziyannis, R. Gish, J. Fry, C. Brosgart, C. Gibbs, M. Miller, and S. Xiong. 2003. Hepatitis B virus genotypes and virologic response in 694 patients in phase III studies of adefovir dipivoxil1. Gastroenterology 125:107-116. [DOI] [PubMed] [Google Scholar]
- 32.Zeng, G., Z. Wang, S. Wen, J. Jiang, L. Wang, J. Cheng, D. Tan, F. Xiao, S. Ma, W. Li, K. Luo, N. V. Naoumov, and J. Hou. 2005. Geographic distribution, virologic and clinical characteristics of hepatitis B virus genotypes in China. J. Viral Hepat. 12:609-617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.