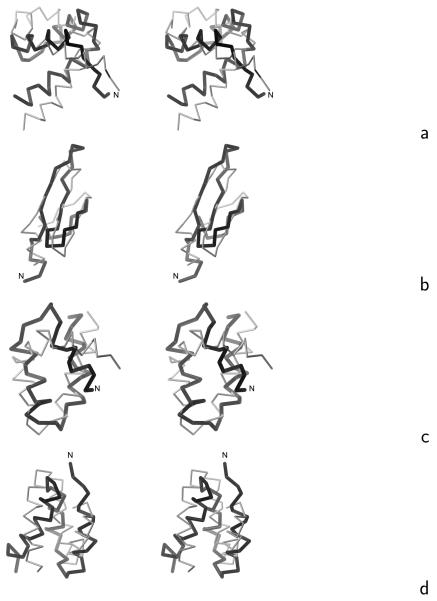
Fig. 6.
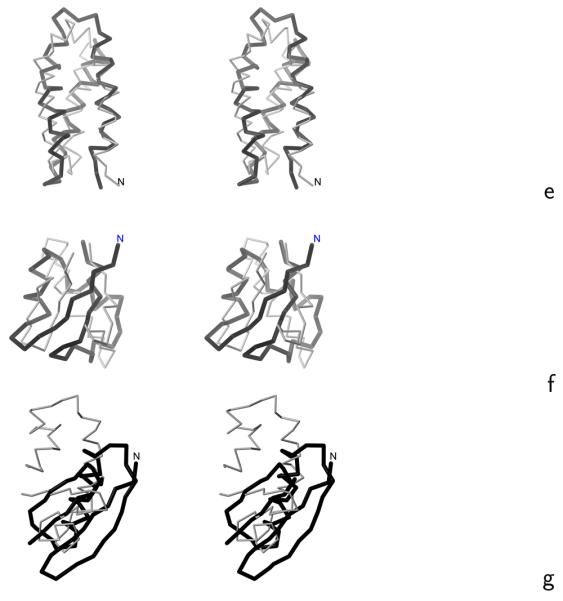
Stereoscopic views of the Cα traces of the average structures of the most native-like clusters of the proteins studied obtained in MREMD simulations with the UNRES force field incorporating the physics-based Ub potentials introduced in ref. 4 and the physics-based Urot and Ubond potentials introduced in this work (grey thin sticks) superposed on the Cα traces of the corresponding experimental structures (black thick sticks): (a) 1ENH, (b) 1E0L, (c) 1BDD, (d) 1GAB, (e) 1LQ7, (f) 1E0G, (g) 1PGA. For 1PGA only the fragments encompassing the middle helix and C-terminal β-hairpin (from residues 20 to 56) are superposed. The RMSD's between the computed and the experimental structures are listed in the ρave column of Table 4. The drawings were done with MOLMOL.29