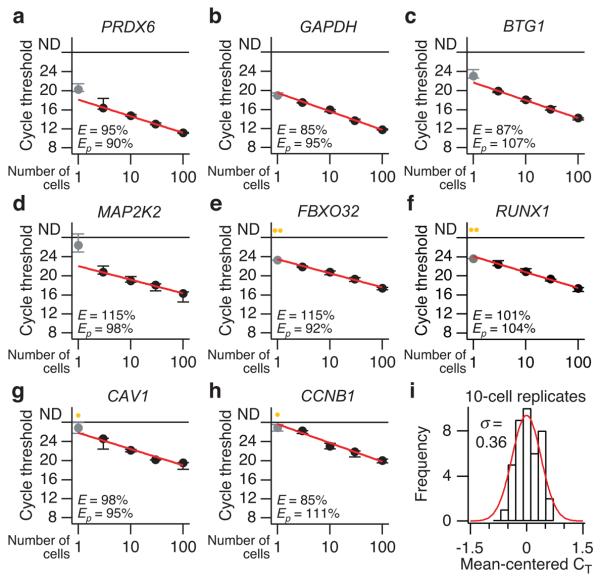
Figure 2. Quantitative and reproducible small-cell amplification of high- to low-abundance transcripts from 3–100 cells.
(a–h) The RT-qPCR cycle threshold for each gene is plotted as a function of starting cellular material and is shown as the median ± range of three replicate small-cell amplifications. Amplification efficiencies (E) based on a log-linear fit of the 3–100-cell dilutions (red line) are listed along with primer efficiencies (Ep) calculated by serially diluting the template before RT-qPCR. Genes are ordered a through h in the order of increasing median cycle threshold from the 10-cell replicates, which was used as an approximation of relative abundance (lower cycle thresholds suggest increased relative abundance). Note that the one-cell amplifications (gray) of higher-abundance transcripts (a-d) often deviate from the log-linear fit, and the one-cell amplification of lower-abundance transcripts (e-h) are frequently not detectable (yellow, ND). (i) Reproducible small-cell amplification of 10 cells. The cycle thresholds from 10-cell amplification replicates of all genes were mean centered, grouped, and fit to a normal distribution. The standard deviation (σ) of the mean-centered cycle thresholds (CT) was 0.36, corresponding to a coefficient of variation of 28%, assuming that amplicons double after each cycle (i.e., 100% efficiency, 20.36 – 1 = 0.28).